bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-32_CDS_annotation_glimmer3.pl_2_4
Length=497
Score E
Sequences producing significant alignments: (Bits) Value
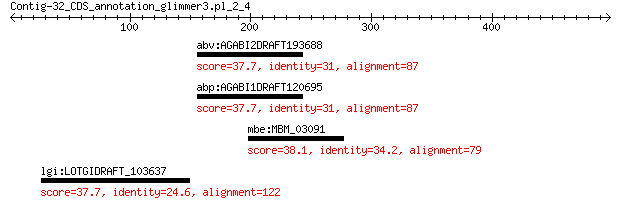
abv:AGABI2DRAFT193688 AGABI2DRAFT_193688; hypothetical protein 37.7 3.7
abp:AGABI1DRAFT120695 AGABI1DRAFT_120695; hypothetical protein 37.7 3.9
mbe:MBM_03091 hypothetical protein 38.1 4.9
lgi:LOTGIDRAFT_103637 hypothetical protein 37.7 6.7
> abv:AGABI2DRAFT193688 AGABI2DRAFT_193688; hypothetical protein
Length=216
Score = 37.7 bits (86), Expect = 3.7, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 44/92 (48%), Gaps = 15/92 (16%)
Query 156 DEILAKKGNQTFYLG----DGKSSTQIANV-FNERGDNQKLKTTRPQYGLLLKTYNSDLY 210
D + +G+Q Y+G DG+++T+ V ++ R D+++L+ P Y LLK +
Sbjct 32 DAVRKAQGSQHIYVGKCVDDGETNTRFVFVTWDSRADHKRLEEDAPVYEQLLKDF----- 86
Query 211 QNWINTEWIDGANGINEITAVDVSDGTLSMDA 242
T+ DGA G N V D T+ A
Sbjct 87 -----TKCADGAAGFNVFHIVSSDDPTMVFKA 113
> abp:AGABI1DRAFT120695 AGABI1DRAFT_120695; hypothetical protein
Length=216
Score = 37.7 bits (86), Expect = 3.9, Method: Compositional matrix adjust.
Identities = 27/92 (29%), Positives = 43/92 (47%), Gaps = 15/92 (16%)
Query 156 DEILAKKGNQTFYLG----DGKSSTQIANV-FNERGDNQKLKTTRPQYGLLLKTYNSDLY 210
D + +G+Q Y+G DG++ T+ V ++ R D+++L+ P Y LLK +
Sbjct 32 DAVRKAQGSQHIYVGKCVDDGETDTRFVFVTWDSRADHKRLEEDAPVYEQLLKDF----- 86
Query 211 QNWINTEWIDGANGINEITAVDVSDGTLSMDA 242
T+ DGA G N V D T+ A
Sbjct 87 -----TKCADGAAGFNVFHIVSSDDPTMVFKA 113
> mbe:MBM_03091 hypothetical protein
Length=954
Score = 38.1 bits (87), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 27/89 (30%), Positives = 44/89 (49%), Gaps = 10/89 (11%)
Query 198 YGLLLKTYNSDLYQNWINTEWIDGANGINEITAVDV-SDGTLSMDALNLQQK----VYNM 252
+ + +K YN L W TEWI IN +TA+ V S G LS+ A+ L++ +Y+
Sbjct 489 HNIPVKQYNDKLGARWFATEWISLDVAINGVTALGVQSIGPLSLQAMVLREPDLKLIYSR 548
Query 253 LNRIAVSGGS-----YRDWLETVYASGQY 276
+ ++ G S Y +E+ G+Y
Sbjct 549 IAQLRAGGVSIGNSLYSQAVESFAKDGKY 577
> lgi:LOTGIDRAFT_103637 hypothetical protein
Length=580
Score = 37.7 bits (86), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 30/133 (23%), Positives = 60/133 (45%), Gaps = 11/133 (8%)
Query 27 FYANKQEENYYMIGSNDPLSITINNTQITDPNNIPQNQGTVNAKSIIEI----------D 76
F ++Q + Y + N + +ITD + P QG I+E+ +
Sbjct 151 FTMDRQRNSSYRVSRNSDIYRKSPAIKITDLDVSPDLQGLKIGDRILEVNGLNVRDKSVE 210
Query 77 DVLSIYNNSNVTLWVTQQIGHSPVKMTADEIGTTSTDN-QKFIIQNLKIPTGNTWWIRTI 135
D+ +I+NN+N L +T + SP++ ++D T T + + II N + ++
Sbjct 211 DIDTIFNNTNEVLHITLERDPSPLRHSSDSDSPTDTSSPDQVIIGNTPVQLRPRSSLKAR 270
Query 136 QSTQQQALVPFPL 148
+++++ P PL
Sbjct 271 NPSRRRSKSPSPL 283
Lambda K H a alpha
0.314 0.131 0.390 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1084184891959