bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-2_CDS_annotation_glimmer3.pl_2_7
Length=171
Score E
Sequences producing significant alignments: (Bits) Value
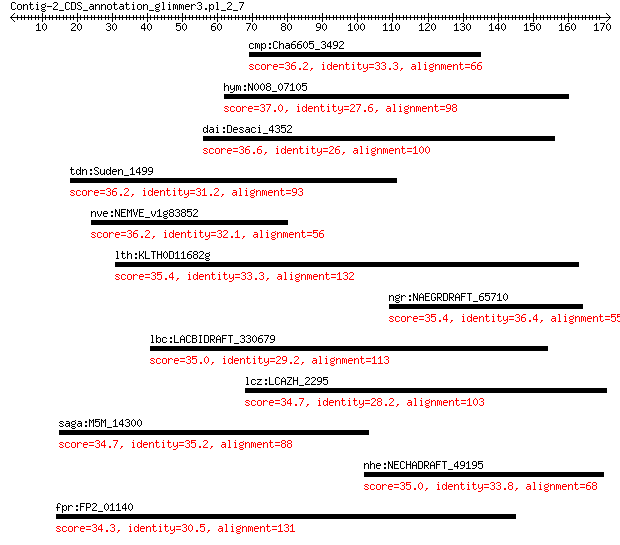
cmp:Cha6605_3492 hypothetical protein 36.2 0.59
hym:N008_07105 hypothetical protein 37.0 1.4
dai:Desaci_4352 YhgE/Pip-like protein 36.6 2.1
tdn:Suden_1499 resistance-nodulation-cell division family tran... 36.2 2.8
nve:NEMVE_v1g83852 hypothetical protein 36.2 3.0
lth:KLTH0D11682g KLTH0D11682p 35.4 5.3
ngr:NAEGRDRAFT_65710 hypothetical protein 35.4 5.7
lbc:LACBIDRAFT_330679 hypothetical protein 35.0 5.8
lcz:LCAZH_2295 BhtR 34.7 6.3
saga:M5M_14300 hypothetical protein 34.7 8.0
nhe:NECHADRAFT_49195 hypothetical protein 35.0 8.3
fpr:FP2_01140 Predicted transcriptional regulator with C-termi... 34.3 9.0
> cmp:Cha6605_3492 hypothetical protein
Length=95
Score = 36.2 bits (82), Expect = 0.59, Method: Compositional matrix adjust.
Identities = 22/67 (33%), Positives = 36/67 (54%), Gaps = 1/67 (1%)
Query 69 HAADLAKRAGVSTVQRFLDSKSPRSSSLQEQLDKLNPSDDELLSMVKSRHLQHPSEI-LA 127
H DLA + TV++ ++ S ++Q+ D LNPS E+ + +S L PSE+ L
Sbjct 12 HILDLAIKHNDKTVKQLVNYPSSLLIAMQQYKDNLNPSYTEIYKIFESGLLLSPSEVDLN 71
Query 128 WIDSINE 134
W+ + N
Sbjct 72 WLKNQNN 78
> hym:N008_07105 hypothetical protein
Length=441
Score = 37.0 bits (84), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 27/99 (27%), Positives = 50/99 (51%), Gaps = 5/99 (5%)
Query 62 SDVSMLLHAADLAKRAGVSTVQRFLDSKSPRSSSLQEQLDKLNPSDDELLSMVKSRHLQH 121
S+VS+ + A KR+GV+T +F D+ R E++ +L P D E + M+ ++
Sbjct 51 SNVSLAVEA-RFGKRSGVATCNQFDDATLRRCVQRAEEIARLAPEDPEYMPMLGAQQYLT 109
Query 122 PSEILAWIDSINELAEDMRSEALKQT-AENESTKFDSSG 159
P+ A S + D R++ + + A E+ K ++G
Sbjct 110 PTTYAA---STAGITPDFRAQVAQDSIALCEARKLTAAG 145
> dai:Desaci_4352 YhgE/Pip-like protein
Length=720
Score = 36.6 bits (83), Expect = 2.1, Method: Composition-based stats.
Identities = 26/105 (25%), Positives = 53/105 (50%), Gaps = 5/105 (5%)
Query 56 VSYRLRSDVSMLLHAADLAKRAG--VSTVQRFLDSKSPRSSSLQEQLDKLN---PSDDEL 110
V+ L+++ + +L D +A V +Q F+D + S SLQ L + P ++
Sbjct 179 VAENLQTNKAKILQLKDTVTQASTNVEAIQGFIDKANSNSESLQGYLKSVQNTLPKINDQ 238
Query 111 LSMVKSRHLQHPSEILAWIDSINELAEDMRSEALKQTAENESTKF 155
+S +++ + S +L+ SIN A D+ ++ ++ A N+ T+
Sbjct 239 ISSLQNATEANKSLVLSTKQSINNTAADLNNDLIQLEAINQQTQL 283
> tdn:Suden_1499 resistance-nodulation-cell division family transporter
Length=1005
Score = 36.2 bits (82), Expect = 2.8, Method: Composition-based stats.
Identities = 29/98 (30%), Positives = 48/98 (49%), Gaps = 6/98 (6%)
Query 18 FTPTREISTFPSTRIGEISLE---VDPVEQFRFETETFGESVSYRLRSDVSMLLHAADLA 74
F ++ +TF + EI E D ++ F + FGE S +L + V+ ++ D
Sbjct 728 FLEQKQSTTFNERGVMEIKTEDIKKDSIDTFLNFSIPFGEGKSVKL-TQVADIIEIRDYE 786
Query 75 K--RAGVSTVQRFLDSKSPRSSSLQEQLDKLNPSDDEL 110
K + S ++ F + R ++ QE LDKL P+ DEL
Sbjct 787 KINKLNGSIIKTFFATIDKRKTTSQEVLDKLEPTLDEL 824
> nve:NEMVE_v1g83852 hypothetical protein
Length=796
Score = 36.2 bits (82), Expect = 3.0, Method: Composition-based stats.
Identities = 18/57 (32%), Positives = 31/57 (54%), Gaps = 1/57 (2%)
Query 24 ISTFPSTRIGEISLEVDPVEQFRFETETFGESV-SYRLRSDVSMLLHAADLAKRAGV 79
+S P + L+ DPVE F T+TF ++ SY +++ SM LH ++ + G+
Sbjct 627 VSVLPGKAVQPADLDYDPVEDAIFWTDTFTGTINSYNMQTRKSMTLHRCNVERPDGI 683
> lth:KLTH0D11682g KLTH0D11682p
Length=1500
Score = 35.4 bits (80), Expect = 5.3, Method: Compositional matrix adjust.
Identities = 44/168 (26%), Positives = 61/168 (36%), Gaps = 38/168 (23%)
Query 31 RIGEISLEVDPVEQF---------------RFETETFGESVSYRLRSDVSMLLHAADLAK 75
RIG LE+D V QF FE+ G + + S ++ L K
Sbjct 261 RIGNSLLEIDAVPQFINNRNEIDFRDIHGTLFESSQIGPDETLFIPSTKNLWTQTTHLRK 320
Query 76 RAGVSTVQ------RFLDSKSPRSSSLQEQLDKLNPSDDELLSMVKS------------- 116
+ G+ Q RF S+ P S +E L L + D + KS
Sbjct 321 QFGLDPYQPIPEQHRF--SRLPEWQSHKELLSFLQKAKDRTIPKSKSFSESVSKSMFSAA 378
Query 117 RHLQHPSEILA--WIDSINELAEDMRSEALKQTAENESTKFDSSGSDV 162
RHL P E L W DS + D+ Q ES DS+ +D+
Sbjct 379 RHLDLPHESLCQLWPDSHPRVGLDLSHRVQNQNTNRESFNSDSTSNDI 426
> ngr:NAEGRDRAFT_65710 hypothetical protein
Length=791
Score = 35.4 bits (80), Expect = 5.7, Method: Composition-based stats.
Identities = 20/55 (36%), Positives = 33/55 (60%), Gaps = 1/55 (2%)
Query 109 ELLSMVKSRHLQHPSEILAWIDSINELAEDMRSEALKQTAENESTKFDSSGSDVS 163
E +S +KS + EI I+ IN++ ED+RS+ ++ +NE KFD+ DV+
Sbjct 186 ETVSEIKSLIINPSKEICVSIEGINDILEDVRSK-VQDLEDNELPKFDTLHDDVA 239
> lbc:LACBIDRAFT_330679 hypothetical protein
Length=352
Score = 35.0 bits (79), Expect = 5.8, Method: Compositional matrix adjust.
Identities = 33/115 (29%), Positives = 59/115 (51%), Gaps = 9/115 (8%)
Query 41 PVEQFRFETET-FGESVSYRLRSDVSMLLHAAD-LAKRAGVSTVQRFLDSKSPRSSSLQE 98
P+E R T FG++++Y + + +H L R ST LDS +S L E
Sbjct 214 PIESTRAIIPTAFGQAMAY---APYPIFIHTHRRLGPRPPRSTKLVELDS----NSRLWE 266
Query 99 QLDKLNPSDDELLSMVKSRHLQHPSEILAWIDSINELAEDMRSEALKQTAENEST 153
++ +PS D +L ++++ QH S+ L D+++ L+ + EA++ EN +T
Sbjct 267 RVTDPDPSPDPVLQDMQAKLDQHDSQELTHDDAMDSLSGRIHQEAVRSLPENITT 321
> lcz:LCAZH_2295 BhtR
Length=216
Score = 34.7 bits (78), Expect = 6.3, Method: Compositional matrix adjust.
Identities = 29/108 (27%), Positives = 54/108 (50%), Gaps = 5/108 (5%)
Query 68 LHAADLAKRAGVSTVQRFLDSKSPRSS-SLQEQLDKLNPSDDELLSMV-KSRHLQHPSEI 125
L AD+A VSTV +F + S S+ L + L+KL E ++ S+H +H S
Sbjct 19 LRIADVAGEISVSTVSKFENGHSEISAEKLMQLLNKLGMDATEFFEILDYSQHQKHISTN 78
Query 126 LA---WIDSINELAEDMRSEALKQTAENESTKFDSSGSDVSGAESVPV 170
L+ +I ++N+L + + LK+ + +F+ + S + + + V
Sbjct 79 LSQKGFIKTLNQLGLEQDIDGLKKFRLRFTKQFNKTQSRLCKLQEIIV 126
> saga:M5M_14300 hypothetical protein
Length=244
Score = 34.7 bits (78), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 31/90 (34%), Positives = 43/90 (48%), Gaps = 9/90 (10%)
Query 15 FREFTPTREISTFPSTRIGEISLEVDPVEQFRFETETFGESVSYRLRSDVSMLLHAADLA 74
F I+ +R+ E+ L + V ET Y L S S A A
Sbjct 87 LNRFPAASGINNHMGSRLTEMVLPMRAV------METLAPRRLYFLDSKTSRRSVAWQEA 140
Query 75 KRAGVSTVQR--FLDSKSPRSSSLQEQLDK 102
KRAG+ TVQR FLD++ P ++++Q QLDK
Sbjct 141 KRAGLETVQRDVFLDNE-PSAAAIQVQLDK 169
> nhe:NECHADRAFT_49195 hypothetical protein
Length=764
Score = 35.0 bits (79), Expect = 8.3, Method: Compositional matrix adjust.
Identities = 23/68 (34%), Positives = 35/68 (51%), Gaps = 13/68 (19%)
Query 102 KLNPSDDELLSMVKSRHLQHPSEILAWIDSINELAEDMRSEALKQTAENESTKFDSSGSD 161
K P+ +ELL M+K H W +E+A+D+R +K+TA+ +T G+
Sbjct 568 KTTPTKEELLGMIKRSH-------FIW----SEVAQDVRE--VKKTADTLATMLAKLGAP 614
Query 162 VSGAESVP 169
V G SVP
Sbjct 615 VQGPASVP 622
> fpr:FP2_01140 Predicted transcriptional regulator with C-terminal
CBS domains
Length=184
Score = 34.3 bits (77), Expect = 9.0, Method: Compositional matrix adjust.
Identities = 40/141 (28%), Positives = 63/141 (45%), Gaps = 17/141 (12%)
Query 14 GFREFTPTREISTFPSTRIGEISLEVDPVEQFRFETETFGESVSYRLRSDVSMLLHAADL 73
GFR T ++ ++ +LE P ++ E+ S + R V +L+ AD
Sbjct 39 GFRVLTQSK-------IKVIADALETTPSYIMGWDEESHQNEWSSKFRDSVMQILNNADP 91
Query 74 A--KRAGVST--VQRFLDSKSPRSSSLQEQL-DKLNPSDDELLS-----MVKSRHLQHPS 123
A K AG+S ++ LD P S + + D+L S D LL M+K+ Q
Sbjct 92 ADLKAAGISVQEIEEELDGSEPISLAAACSIADQLGESLDSLLGHTPKEMIKAALQQEDG 151
Query 124 EILAWIDSINELAEDMRSEAL 144
+ ID + +L+ D R EAL
Sbjct 152 QTAEIIDLLLDLSADRRQEAL 172
Lambda K H a alpha
0.312 0.127 0.347 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 137967995676