bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-29_CDS_annotation_glimmer3.pl_2_1
Length=325
Score E
Sequences producing significant alignments: (Bits) Value
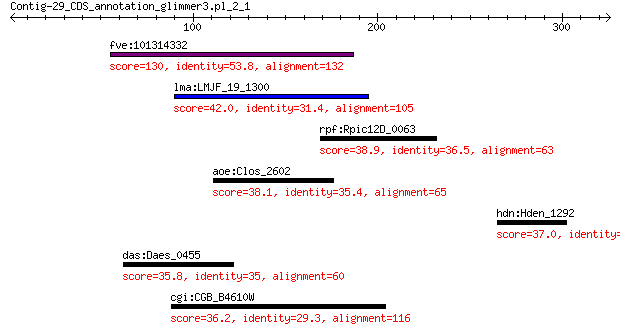
fve:101314332 capsid protein VP1-like 130 3e-31
lma:LMJF_19_1300 hypothetical protein 42.0 0.19
rpf:Rpic12D_0063 hypothetical protein 38.9 1.5
aoe:Clos_2602 S-layer domain-containing protein 38.1 2.2
hdn:Hden_1292 3-oxoacyl-(acyl-carrier-protein) synthase III (E... 37.0 5.0
das:Daes_0455 hypothetical protein 35.8 5.7
cgi:CGB_B4610W hypothetical protein 36.2 6.9
> fve:101314332 capsid protein VP1-like
Length=421
Score = 130 bits (326), Expect = 3e-31, Method: Compositional matrix adjust.
Identities = 71/134 (53%), Positives = 89/134 (66%), Gaps = 3/134 (2%)
Query 55 SAVTPINLWAVNDGSVSSATINQLRLAFQVQKLYERDARGGTRYIEVLKSHFGVTSPDAR 114
SA L+A + + ++ATINQLR +FQ+QKL ERDARGGTRY E+++SHFGV SPDAR
Sbjct 289 SATVATGLYA-DLSAATAATINQLRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDAR 347
Query 115 LQRPEYLGGNRIPIVISEI--NQTSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVEHGFI 172
LQRPEYLGG PI I+ I +G +TPQGN + F +SFVEHG +
Sbjct 348 LQRPEYLGGGSTPINIAPIAQTGGTGAQGTTTPQGNLAAFGTYMAKGHGFSQSFVEHGHV 407
Query 173 IGVMVARYDHTYQQ 186
IG++ R D TYQQ
Sbjct 408 IGLVSVRADLTYQQ 421
> lma:LMJF_19_1300 hypothetical protein
Length=851
Score = 42.0 bits (97), Expect = 0.19, Method: Compositional matrix adjust.
Identities = 33/118 (28%), Positives = 47/118 (40%), Gaps = 15/118 (13%)
Query 90 RDARGGTRYIEVLKSH------FGVTSPDARLQRPEYLGGNRIPIVISEINQTSGTSANS 143
RD R G VL++H GV + + R+ R Y GG PIV + + Q +G A
Sbjct 216 RDGRDGVLGRRVLEAHPNVVTLLGVVTAEERVCRCRYAGGIETPIVTAAMMQGAGPVAGG 275
Query 144 TPQGNPSGQSRTTDVH-------SDFKKSFVEHGFIIGVMVARYDHTYQQGLERFWSR 194
P Q R + DF ++ + F+ V V R+ GL SR
Sbjct 276 DPAAASLCQVRAVLLENVSGGAWCDFLATYGK--FVTPVCVMRWFRDVVTGLAHLHSR 331
> rpf:Rpic12D_0063 hypothetical protein
Length=862
Score = 38.9 bits (89), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 23/63 (37%), Positives = 35/63 (56%), Gaps = 1/63 (2%)
Query 169 HGFIIGVMVARYDHTYQQGLERFWSRKGRLDYYWPVFANIGEQAVLNKEIYAQGNGTDDE 228
H F+ VM A D + + +R W+RKGRL + IG AV ++E +A+GN T
Sbjct 717 HPFVARVMRAMPDADWAERGQR-WARKGRLVGFLGALVGIGWDAVHSREEFAKGNQTLGY 775
Query 229 VFG 231
++G
Sbjct 776 LYG 778
> aoe:Clos_2602 S-layer domain-containing protein
Length=493
Score = 38.1 bits (87), Expect = 2.2, Method: Compositional matrix adjust.
Identities = 23/67 (34%), Positives = 35/67 (52%), Gaps = 2/67 (3%)
Query 111 PDARLQRPEYLGG--NRIPIVISEINQTSGTSANSTPQGNPSGQSRTTDVHSDFKKSFVE 168
PD + R E+ I ++E+ +T G+S N TP+ +P TTD + D+ K+ V
Sbjct 319 PDVPMNREEFTRAVMRASAIRLAEVPKTRGSSRNKTPEISPFSDVATTDANYDYIKNGVA 378
Query 169 HGFIIGV 175
G I GV
Sbjct 379 KGIIQGV 385
> hdn:Hden_1292 3-oxoacyl-(acyl-carrier-protein) synthase III
(EC:2.3.1.41)
Length=326
Score = 37.0 bits (84), Expect = 5.0, Method: Compositional matrix adjust.
Identities = 17/37 (46%), Positives = 24/37 (65%), Gaps = 0/37 (0%)
Query 265 DDYSKLPSLSDSWVQEDSAVVNRVIAVSEENSNQLWA 301
+D SK+ SD+W+QE S +VNR IA E ++ L A
Sbjct 24 EDLSKIVDTSDAWIQERSGIVNRHIADETEKTSDLGA 60
> das:Daes_0455 hypothetical protein
Length=150
Score = 35.8 bits (81), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 21/61 (34%), Positives = 32/61 (52%), Gaps = 3/61 (5%)
Query 62 LWAVNDGSVSSATINQLRLAFQVQKLYERDARGGTRYIEVLKSHFGVTSPD-ARLQRPEY 120
L+ N GS + AF+++ L + D++GG ++I K F VTSPD R+ Y
Sbjct 83 LFYANSGSGEELKVEAFGCAFKIESLLDIDSKGGEKHIT--KLPFDVTSPDFIRINGVPY 140
Query 121 L 121
L
Sbjct 141 L 141
> cgi:CGB_B4610W hypothetical protein
Length=262
Score = 36.2 bits (82), Expect = 6.9, Method: Compositional matrix adjust.
Identities = 34/131 (26%), Positives = 52/131 (40%), Gaps = 42/131 (32%)
Query 88 YERDARGGTRYIEVLKSHFGVTSPDARLQRPEYLGGNRIPIVISEINQTSGTSANSTPQG 147
+ERD + G+ Y L H + S + RL PEY GN P +S +
Sbjct 110 HERDVQKGSYYANPLMDH-PIVSDETRLAYPEYYAGNIWPKGMSGLE------------- 155
Query 148 NPSGQSRTTDVHSDFKKSFVEHG---FIIGVMVARY------------DHTYQQGLERFW 192
DF+++F E G F +GV++AR + T + +
Sbjct 156 -------------DFEQTFKELGKLIFDVGVLLARVCDNFVTPTLANPEGTLSSLIAKSK 202
Query 193 SRKGRLDYYWP 203
S K RL +Y+P
Sbjct 203 SSKARLLHYYP 213
Lambda K H a alpha
0.315 0.132 0.402 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 578736825155