bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-24_CDS_annotation_glimmer3.pl_2_5
Length=421
Score E
Sequences producing significant alignments: (Bits) Value
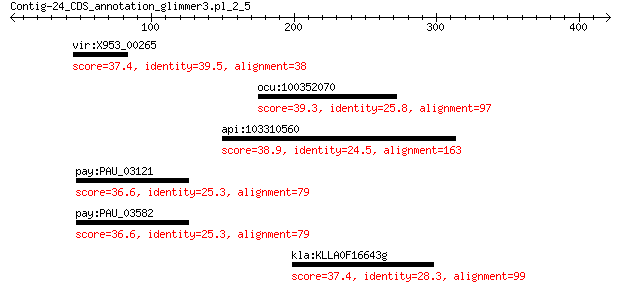
vir:X953_00265 hypothetical protein 37.4 1.1
ocu:100352070 AASS; aminoadipate-semialdehyde synthase 39.3 1.7
api:103310560 centromere-associated protein E-like 38.9 2.4
pay:PAU_03121 gene0067; similar to cryptic prophage endopeptid... 36.6 3.4
pay:PAU_03582 gene0098; similar to cryptic prophage endopeptid... 36.6 4.7
kla:KLLA0F16643g hypothetical protein 37.4 6.8
> vir:X953_00265 hypothetical protein
Length=97
Score = 37.4 bits (85), Expect = 1.1, Method: Composition-based stats.
Identities = 15/40 (38%), Positives = 30/40 (75%), Gaps = 2/40 (5%)
Query 45 KYKQKEMALQQQYALEQMSKSAEFQLAHD--KQMFDYQNS 82
K+ K+ A+Q++Y ++Q+S++ +FQL D K++ +Y+NS
Sbjct 55 KFPTKQEAMQKEYQIKQLSRTEKFQLIRDRLKEVMEYENS 94
> ocu:100352070 AASS; aminoadipate-semialdehyde synthase
Length=926
Score = 39.3 bits (90), Expect = 1.7, Method: Compositional matrix adjust.
Identities = 25/97 (26%), Positives = 48/97 (49%), Gaps = 1/97 (1%)
Query 175 RSQTLDKDLRERLMKAQAGLAEAGITESASRASLNAAITLSYSIDNELKDAAFGYNLEMI 234
R+Q+L D +++++ +G + E SR S N IT+ + N+++ + YN+ +
Sbjct 471 RAQSLSMDTKKKVLVLGSGYVSEPVLEYLSRDS-NIEITVGSDMKNQIEQLSKKYNITPV 529
Query 235 KANLGKAKEEYYQLKARTGYIDELIEKELQLLTVRAI 271
N+GK +E+ L A + L+ L L +A
Sbjct 530 SMNIGKQEEKLDSLVATQDLVISLLPYVLHPLVAKAC 566
> api:103310560 centromere-associated protein E-like
Length=1861
Score = 38.9 bits (89), Expect = 2.4, Method: Composition-based stats.
Identities = 40/169 (24%), Positives = 73/169 (43%), Gaps = 27/169 (16%)
Query 150 VQLKDAQQERERSAASLNDAEADWYRSQTLDKDLRERLMKAQAGLAEAGITESASRASLN 209
+++KDA +++ A LN+AE D +S ++L+ L + Q L A +
Sbjct 1701 IEIKDANSVKQKLEACLNEAEYDLIKSGDFCENLKFELYRVQRELENACVN--------- 1751
Query 210 AAITLSYSIDNELKDAAFGYNLEMIKANLGKAKEEYYQLKARTGYIDELIEK----ELQL 265
+ S++ EL++ F N E K L + R ID L++ +L+L
Sbjct 1752 -----AESVEKELQNLKFKLNDENDKNELNGS-------DVRNKLIDPLVDYVHDIKLKL 1799
Query 266 LTVRAIYLKSSASNQEQLARVNELTADDL--ENWFDVNWNTQVDVPIID 312
+ + + + S + +V D L E+ +N ++Q D P ID
Sbjct 1800 TELNSAIISGNQSEKRLRTKVLSTEEDILPHEDTLKINCDSQPDSPDID 1848
> pay:PAU_03121 gene0067; similar to cryptic prophage endopeptidase
b1362
Length=149
Score = 36.6 bits (83), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 39/79 (49%), Gaps = 1/79 (1%)
Query 47 KQKEMALQQQYALEQMSKSAEFQLAHDKQMFDYQNSYNDPAAVLERNFSAGLNPAAVLGQ 106
K++ + QQQ ++Q++ + +Q +H + + N + A + GL G+
Sbjct 29 KERHVTKQQQEDIQQLTDTISYQNSHITMLAELDNKHTKELANAKSEID-GLRDDVAAGR 87
Query 107 SGVGVSATIPTSSGGAPSG 125
+ ++AT P S G+PSG
Sbjct 88 RRLRIAATCPKSEAGSPSG 106
> pay:PAU_03582 gene0098; similar to cryptic prophage endopeptidase
b1362
Length=174
Score = 36.6 bits (83), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 20/79 (25%), Positives = 39/79 (49%), Gaps = 1/79 (1%)
Query 47 KQKEMALQQQYALEQMSKSAEFQLAHDKQMFDYQNSYNDPAAVLERNFSAGLNPAAVLGQ 106
K++ + QQQ ++Q++ + +Q +H + + N + A + GL G+
Sbjct 54 KERHVTKQQQEDIQQLTDTISYQNSHITMLAELDNKHTKELANAKSEID-GLRDDVAAGR 112
Query 107 SGVGVSATIPTSSGGAPSG 125
+ ++AT P S G+PSG
Sbjct 113 RRLRIAATCPKSEAGSPSG 131
> kla:KLLA0F16643g hypothetical protein
Length=665
Score = 37.4 bits (85), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 28/114 (25%), Positives = 57/114 (50%), Gaps = 15/114 (13%)
Query 199 ITESASRASLNAAITLSYSIDNELK-----------DAAFGYNLEMIKANLGKAKEEYYQ 247
I ES SR+S+ ++TL +ID+ L+ GY + +++ +L K + ++ +
Sbjct 522 IKESTSRSSMQDSLTLGITIDDNLRAIGSVKAVLNQRKKLGYYILLVENDLNKKQTQFNK 581
Query 248 LKARTGYIDEL--IEKELQLLTVRAIYLKSSASNQEQLAR--VNELTADDLENW 297
+ + ++ IE EL++LT R +K ++ R VN+ D +E++
Sbjct 582 VSGNSANSEKATSIENELRILTSRCNKIKQEWQTIAEVIRKDVNQHDVDTIEDF 635
Lambda K H a alpha
0.312 0.127 0.354 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 864851519007