bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_8
Length=286
Score E
Sequences producing significant alignments: (Bits) Value
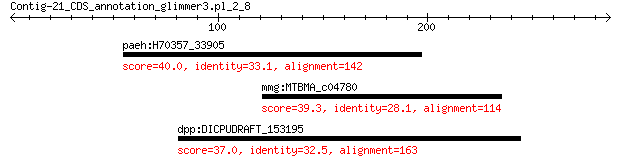
paeh:H70357_33905 response regulator receiver protein 40.0 0.51
mmg:MTBMA_c04780 hypothetical protein 39.3 0.97
dpp:DICPUDRAFT_153195 hypothetical protein 37.0 5.1
> paeh:H70357_33905 response regulator receiver protein
Length=1145
Score = 40.0 bits (92), Expect = 0.51, Method: Compositional matrix adjust.
Identities = 47/148 (32%), Positives = 74/148 (50%), Gaps = 19/148 (13%)
Query 55 IAERQRMKNEKAIADATVNKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQL 114
+ E Q K E +A +L A+ ++L T++ D R + + ++ Q E++Q+
Sbjct 286 LGETQLQKEE---LEAQTEELQAQTQELEAQTEELLAQTDELRSQTEQ--LQAQGEELQM 340
Query 115 AVDEI---NKEFQRAINEADVQIKHGLYSETLAKIDKLIADKDVSEEM---KQNLQKQRD 168
D + NK+ QR + + Q SE A+ D+L K +EE+ Q+LQKQ +
Sbjct 341 QADSLLTSNKQLQRQMELTEEQ-----KSEIEAQADEL---KLQTEELFASNQDLQKQME 392
Query 169 LIESQISATQAQTDLIKAQTSATQAQTE 196
+ E Q QAQ D + AQT QAQTE
Sbjct 393 ISEQQKEEIQAQADELLAQTEELQAQTE 420
> mmg:MTBMA_c04780 hypothetical protein
Length=653
Score = 39.3 bits (90), Expect = 0.97, Method: Compositional matrix adjust.
Identities = 32/120 (27%), Positives = 62/120 (52%), Gaps = 10/120 (8%)
Query 121 KEFQRAINEADVQIKHGLYSETLAKIDKLIADKDVSEEMKQNLQKQRDLIES---QISAT 177
+E+ NEA + ++ E L K+ + +K+V E+ ++ L+ QR+ ++ I +
Sbjct 260 REYFETFNEAADEFFSSIFREELRKVHEAEWEKEV-EKFRKRLRIQRETLQKFQDTIDTS 318
Query 178 QAQTDLIKAQTSATQAQTETENALRDGRIKLTEREANKILADI---GLSEARSLNEYESL 234
+ DL+ A +A + T +RD R K + +E KI+AD G+ EA+ + E + +
Sbjct 319 TRKGDLLYAHYAAVEDVLRT---IRDAREKYSWKEIRKIIADARSKGMVEAQMIQEIDGM 375
> dpp:DICPUDRAFT_153195 hypothetical protein
Length=662
Score = 37.0 bits (84), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 53/190 (28%), Positives = 84/190 (44%), Gaps = 40/190 (21%)
Query 81 KLRGDTKDPNVTK--DSQRLEFDWNLV------KKQREQVQLAVDEINKEFQRAINE--- 129
KL GD K ++ K + Q E +++ K++ E++ + DE K F+ I +
Sbjct 314 KLNGDEKAEDIVKLLEKQYHEMAKSIINENTKLKEELEKINVNKDEYIKSFEDFIKDRYK 373
Query 130 ----ADVQIKHGLYSETLAKIDKLIADKDVSEE---MKQNLQKQRDLIESQISATQAQTD 182
A V + HG E L +DK I + +V E +K L++Q+ E+ I+ Q +
Sbjct 374 VALDATVDLLHGDIGEKLKDMDKYIQE-NVGENQKLVKDTLERQK---ENLINIYNKQAE 429
Query 183 LIK-------AQTSATQAQTETENALRDGRI--KLTEREANKILADIGLSEARSLNEYES 233
IK A T TQ + E LRD KL +R N + L++Y+
Sbjct 430 AIKKSEVEKRAVTQLTQLIVDIEKLLRDASAQGKLVQRSFNNL---------TDLSQYDE 480
Query 234 LIKAMTGTQP 243
LIK + T P
Sbjct 481 LIKELLNTLP 490
Lambda K H a alpha
0.310 0.126 0.336 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 467204637764