bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_3
Length=90
Score E
Sequences producing significant alignments: (Bits) Value
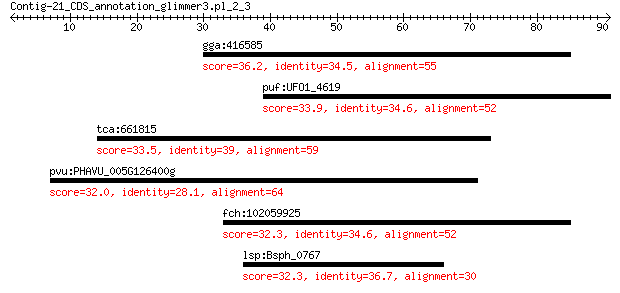
gga:416585 IL4R; interleukin 4 receptor 36.2 0.52
puf:UFO1_4619 Microsomal epoxide hydrolase (EC:3.3.2.9) 33.9 2.8
tca:661815 vacuolar protein sorting-associated protein 54 33.5
pvu:PHAVU_005G126400g hypothetical protein 32.0 5.9
fch:102059925 IL4R; interleukin 4 receptor 32.3 8.8
lsp:Bsph_0767 hypothetical protein 32.3 9.3
> gga:416585 IL4R; interleukin 4 receptor
Length=1112
Score = 36.2 bits (82), Expect = 0.52, Method: Composition-based stats.
Identities = 19/65 (29%), Positives = 31/65 (48%), Gaps = 10/65 (15%)
Query 30 VLVIQYDHFDDFARWKDWVSHVSFPYAYTRAIDDVVTVEVPVE----------PFFRFTT 79
V++I+ ++ D A W DW V F Y Y +D++ + VP+ +F FT
Sbjct 315 VVIIRCNYTDYPAYWSDWSDGVEFHYDYQVTPEDILHMAVPISCILIMAVAVICYFCFTK 374
Query 80 SPKKW 84
K+W
Sbjct 375 MKKEW 379
> puf:UFO1_4619 Microsomal epoxide hydrolase (EC:3.3.2.9)
Length=384
Score = 33.9 bits (76), Expect = 2.8, Method: Compositional matrix adjust.
Identities = 18/52 (35%), Positives = 26/52 (50%), Gaps = 0/52 (0%)
Query 39 DDFARWKDWVSHVSFPYAYTRAIDDVVTVEVPVEPFFRFTTSPKKWHVNNLR 90
D F R+K +S ++ P Y +D V VP P F F++ P +NN R
Sbjct 103 DSFIRYKKIISLLTDPARYGGDPEDSFDVIVPSIPGFGFSSRPDHSGINNSR 154
> tca:661815 vacuolar protein sorting-associated protein 54
Length=818
Score = 33.5 bits (75), Expect = 4.3, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 31/69 (45%), Gaps = 10/69 (14%)
Query 14 TWSDSVTWKTKAGKMPVLV-IQYDHFDDFAR--WKDWVSHVSFP-------YAYTRAIDD 63
TW D+ KT+ K P L I Y HFD + R K + H+ + TR D
Sbjct 39 TWGDTFVEKTQIEKSPFLPEITYSHFDGYVRKIGKRYKRHIRLSQSKLEANHVKTRTKAD 98
Query 64 VVTVEVPVE 72
++ EV VE
Sbjct 99 LINGEVSVE 107
> pvu:PHAVU_005G126400g hypothetical protein
Length=108
Score = 32.0 bits (71), Expect = 5.9, Method: Compositional matrix adjust.
Identities = 18/75 (24%), Positives = 39/75 (52%), Gaps = 11/75 (15%)
Query 7 RGLEALITWSDSV---TWKTKAGKMPVL--------VIQYDHFDDFARWKDWVSHVSFPY 55
+G+E L++ D+V W + VL ++ ++ +DFA ++ +HV F
Sbjct 26 KGMEKLVSEIDAVKAFEWGRDIESLDVLRQGFTHAFLMTFNKKEDFAAFQSHPNHVEFST 85
Query 56 AYTRAIDDVVTVEVP 70
++ AI+++V ++ P
Sbjct 86 TFSAAIENIVLLDFP 100
> fch:102059925 IL4R; interleukin 4 receptor
Length=866
Score = 32.3 bits (72), Expect = 8.8, Method: Composition-based stats.
Identities = 18/62 (29%), Positives = 28/62 (45%), Gaps = 10/62 (16%)
Query 33 IQYDHFDDFARWKDWVSHVSFPYAYTRAIDDVVTVEVPVE----------PFFRFTTSPK 82
++ ++ D A W +W V F Y Y +DV+ + VPV +F FT K
Sbjct 202 VRCNYTDYPAYWSEWSDKVEFHYDYQVTTEDVLQMAVPVSCILIMAVSVTCYFCFTKVKK 261
Query 83 KW 84
+W
Sbjct 262 EW 263
> lsp:Bsph_0767 hypothetical protein
Length=335
Score = 32.3 bits (72), Expect = 9.3, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 18/31 (58%), Gaps = 1/31 (3%)
Query 36 DHFDDFARWKDWVSHV-SFPYAYTRAIDDVV 65
D +D RWK+W++H+ FP+ +D V
Sbjct 81 DRQEDLIRWKEWIAHIFPFPFCKVTVTNDAV 111
Lambda K H a alpha
0.322 0.134 0.459 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 128026330890