bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_6
Length=367
Score E
Sequences producing significant alignments: (Bits) Value
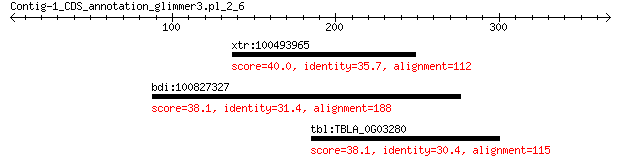
xtr:100493965 rock2; Rho-associated, coiled-coil containing pr... 40.0 0.94
bdi:100827327 uncharacterized LOC100827327 38.1 3.3
tbl:TBLA_0G03280 TBLA0G03280; hypothetical protein 38.1 3.4
> xtr:100493965 rock2; Rho-associated, coiled-coil containing
protein kinase 2
Length=1373
Score = 40.0 bits (92), Expect = 0.94, Method: Compositional matrix adjust.
Identities = 40/119 (34%), Positives = 57/119 (48%), Gaps = 14/119 (12%)
Query 137 LSALQAEANIELTKSQALKTRAETTG-----LENTNS--MFDVVKSIADEELTSKRFSNI 189
L ALQ E + +A K R +E T S M D+ K + +E L +R N
Sbjct 681 LKALQQSVEKEEAEHKATKARLADKNKIYQSIEETKSEAMKDMEKKLHEERLVKQRLENS 740
Query 190 LKEVEVKYAEVNAITDLDAKQAKIAEINASALERLASAAKTDADRITVELLRDAQKRSL 248
L E E +Y+ + D D KQAK +IN LE L D +T++ ++AQKR+L
Sbjct 741 LLETEKQYS----MLDCDLKQAK-QKINE--LESLKDKLSEDVKNLTLKAEQEAQKRNL 792
> bdi:100827327 uncharacterized LOC100827327
Length=476
Score = 38.1 bits (87), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 59/215 (27%), Positives = 86/215 (40%), Gaps = 34/215 (16%)
Query 88 YADVGGIQTSSVDP-------GSYGGSTPSAQSFTQPGGIPSSPLVGAFGNATQQTLSAL 140
+A G SS P G S P+ + P PS P G + + A
Sbjct 82 HASATGNDDSSTRPRKVKLKIGGISRSIPAKPNPDAPDSRPSQP-----GESRHRQKHAS 136
Query 141 QAEANIELTKSQALKTRAETTGLENT-----------NSMFDVV--------KSIADEEL 181
Q E + ++SQ KTR E LE+T S D V KSI D EL
Sbjct 137 QTEGTKDSSRSQDKKTRRERA-LEDTFTPEQPAKVQRESSSDPVRKSRRLAKKSILDSEL 195
Query 182 TSKRFSNILKEVEVKYAEVNAITDLDAKQAKIAEINASALERLASAA-KTDADRITVELL 240
+ ++ L+ + I D K ++ NAS +R S + D D +T +
Sbjct 196 DEEYDTSALENLGASNDNDGRIRDSKNKGGSNSKKNASKKDRSRSTVYEVDNDFVTPQSN 255
Query 241 RDAQKRSLEAGASLAEAQAATEPHRALNLKQDTLL 275
RD +KRS E+ + E + +EP A N KQ ++
Sbjct 256 RDGKKRSRESTDAEEELASDSEPE-AENKKQKAVV 289
> tbl:TBLA_0G03280 TBLA0G03280; hypothetical protein
Length=964
Score = 38.1 bits (87), Expect = 3.4, Method: Compositional matrix adjust.
Identities = 35/118 (30%), Positives = 56/118 (47%), Gaps = 8/118 (7%)
Query 185 RFSNILKEVEVKYAEVNAITDLDAKQAKIAEINASALERLASAAKTDADRITVELLRDAQ 244
+ SN LKE + E+ + + + + E NA ALER + A+ A + + +
Sbjct 582 KLSNYLKEENENFKELTYLQKIKMARTNMIEQNARALERKKTIAEERAKKSY-----ERK 636
Query 245 KRSLEAGASLAEAQAATEPHRALNLKQDTLLKMAQEETEQLLRSQKF---ELTRQQAR 299
++ LEA A AE A R LN K + KM +E +LL +K EL ++Q +
Sbjct 637 QKHLEAAAINAEQDAELRQQRMLNEKANIEAKMEEEANRRLLEKKKRAFAELQKEQVK 694
Lambda K H a alpha
0.311 0.123 0.331 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 703779073748