bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_1
Length=164
Score E
Sequences producing significant alignments: (Bits) Value
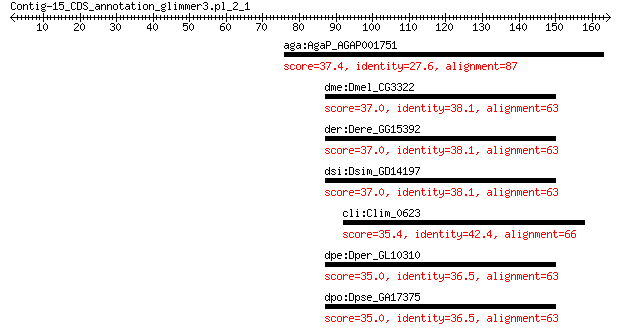
aga:AgaP_AGAP001751 AGAP001751-PE 37.4 1.0
dme:Dmel_CG3322 LanB2; Laminin B2 37.0 1.3
der:Dere_GG15392 Dere_LanB2; Laminin B2 37.0 1.4
dsi:Dsim_GD14197 Dsim_LanB2; GD14197 gene product from transcr... 37.0 1.6
cli:Clim_0623 DNA repair protein RecN 35.4 4.9
dpe:Dper_GL10310 GL10310 gene product from transcript GL10310-RA 35.0 8.0
dpo:Dpse_GA17375 GA17375 gene product from transcript GA17375-RA 35.0 8.0
> aga:AgaP_AGAP001751 AGAP001751-PE
Length=1169
Score = 37.4 bits (85), Expect = 1.0, Method: Compositional matrix adjust.
Identities = 24/87 (28%), Positives = 47/87 (54%), Gaps = 3/87 (3%)
Query 76 SKFGQSKQPISQIQQIMDTMSDEDLLATIRSRYIQSPSEIIAWSKELSAYAENLESQAQE 135
SK G+ K+ + I I T+ D D L ++ +R+ +PSE+ ++ S++ + S Q
Sbjct 137 SKDGEKKRSMPTIPTITYTVKDRDTLTSVAARFDTTPSELTQLNRLASSF---IYSGQQL 193
Query 136 LIDAENARQEAEKAAAAPAAASADAPS 162
L+ +NA+ A+ + A S ++P+
Sbjct 194 LVPDKNAKASADGESDASTERSTNSPT 220
> dme:Dmel_CG3322 LanB2; Laminin B2
Length=1639
Score = 37.0 bits (84), Expect = 1.3, Method: Composition-based stats.
Identities = 24/69 (35%), Positives = 41/69 (59%), Gaps = 6/69 (9%)
Query 87 QIQQIMDTMSDEDLLATIRSR-----YIQSPSEIIAWSKELSAYAENLESQAQ-ELIDAE 140
++QQ +D ++DE A R++ + Q +I S+E A A+ LES+AQ +L +A+
Sbjct 1170 ELQQALDLLNDEGAQALARAKEKSVEFGQQSEQISDISREARALADKLESEAQFDLKNAK 1229
Query 141 NARQEAEKA 149
+A+ EKA
Sbjct 1230 DAKDAVEKA 1238
> der:Dere_GG15392 Dere_LanB2; Laminin B2
Length=1639
Score = 37.0 bits (84), Expect = 1.4, Method: Composition-based stats.
Identities = 24/69 (35%), Positives = 41/69 (59%), Gaps = 6/69 (9%)
Query 87 QIQQIMDTMSDEDLLATIRSR-----YIQSPSEIIAWSKELSAYAENLESQAQ-ELIDAE 140
++QQ +D ++DE A R++ + Q +I S+E A A+ LES+AQ +L +A+
Sbjct 1170 ELQQALDLLNDEGAQALARAKEKSVEFGQQSEQISDISREARALADKLESEAQFDLKNAK 1229
Query 141 NARQEAEKA 149
+A+ EKA
Sbjct 1230 DAKDAVEKA 1238
> dsi:Dsim_GD14197 Dsim_LanB2; GD14197 gene product from transcript
GD14197-RA
Length=1575
Score = 37.0 bits (84), Expect = 1.6, Method: Composition-based stats.
Identities = 24/69 (35%), Positives = 41/69 (59%), Gaps = 6/69 (9%)
Query 87 QIQQIMDTMSDEDLLATIRSR-----YIQSPSEIIAWSKELSAYAENLESQAQ-ELIDAE 140
++QQ +D ++DE A R++ + Q +I S+E A A+ LES+AQ +L +A+
Sbjct 1106 ELQQALDLLNDEGAQALARAKEKSVEFGQQSEQISDISREARALADKLESEAQFDLKNAK 1165
Query 141 NARQEAEKA 149
+A+ EKA
Sbjct 1166 DAKDAVEKA 1174
> cli:Clim_0623 DNA repair protein RecN
Length=568
Score = 35.4 bits (80), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 28/69 (41%), Positives = 40/69 (58%), Gaps = 4/69 (6%)
Query 92 MDTM-SDEDLLATIRSRYIQSPSEIIAWSKELSAYAENLESQAQE--LIDAENARQEAEK 148
+DTM + + LL R +Y ++ SE+I+W EL+A ES A+E LID E EK
Sbjct 297 LDTMRTRQHLLQRTRKKYAKTLSELISWRDELTAALGIEESIAEENSLIDTEIGSLR-EK 355
Query 149 AAAAPAAAS 157
+AA A+ S
Sbjct 356 LSAAAASLS 364
> dpe:Dper_GL10310 GL10310 gene product from transcript GL10310-RA
Length=1637
Score = 35.0 bits (79), Expect = 8.0, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 40/69 (58%), Gaps = 6/69 (9%)
Query 87 QIQQIMDTMSDEDLLATIRSR-----YIQSPSEIIAWSKELSAYAENLESQAQ-ELIDAE 140
++QQ +D ++DE A +++ + Q +I S+E A A+ LES+AQ +L +A+
Sbjct 1168 ELQQALDLLNDEGTQALAKAKDKSVEFGQQSQQISDISREARALADKLESEAQFDLKNAK 1227
Query 141 NARQEAEKA 149
+A EKA
Sbjct 1228 DAMDAVEKA 1236
> dpo:Dpse_GA17375 GA17375 gene product from transcript GA17375-RA
Length=1637
Score = 35.0 bits (79), Expect = 8.0, Method: Composition-based stats.
Identities = 23/69 (33%), Positives = 40/69 (58%), Gaps = 6/69 (9%)
Query 87 QIQQIMDTMSDEDLLATIRSR-----YIQSPSEIIAWSKELSAYAENLESQAQ-ELIDAE 140
++QQ +D ++DE A +++ + Q +I S+E A A+ LES+AQ +L +A+
Sbjct 1168 ELQQALDLLNDEGTQALAKAKDKSVEFGQQSQQISDISREARALADKLESEAQFDLKNAK 1227
Query 141 NARQEAEKA 149
+A EKA
Sbjct 1228 DAMDAVEKA 1236
Lambda K H a alpha
0.309 0.123 0.327 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 126451599574