bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-12_CDS_annotation_glimmer3.pl_2_6
Length=254
Score E
Sequences producing significant alignments: (Bits) Value
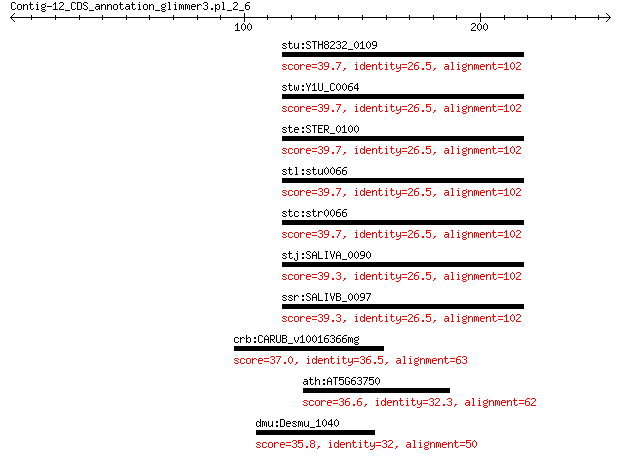
stu:STH8232_0109 hypothetical protein 39.7 0.22
stw:Y1U_C0064 hypothetical protein 39.7 0.24
ste:STER_0100 hypothetical protein 39.7 0.24
stl:stu0066 hypothetical protein 39.7 0.24
stc:str0066 hypothetical protein 39.7 0.24
stj:SALIVA_0090 flavoprotein 39.3 0.30
ssr:SALIVB_0097 Oxidoreductase (EC:1.1.1.-) 39.3 0.36
crb:CARUB_v10016366mg hypothetical protein 37.0 4.8
ath:AT5G63750 ARI13; putative E3 ubiquitin-protein ligase ARI13 36.6 5.4
dmu:Desmu_1040 enolase 35.8 8.6
> stu:STH8232_0109 hypothetical protein
Length=181
Score = 39.7 bits (91), Expect = 0.22, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 42/102 (41%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 80 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 129
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+I + + ND+ W D +F+ VLE
Sbjct 130 KELLPFIRTQVVGDFTATRVNDT----AWVDGKFVVTEEVLE 167
> stw:Y1U_C0064 hypothetical protein
Length=183
Score = 39.7 bits (91), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 42/102 (41%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 80 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 129
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+I + + ND+ W D +F+ VLE
Sbjct 130 KELLPFIRTQVVGDFTATRVNDT----AWVDGKFVVTEEVLE 167
> ste:STER_0100 hypothetical protein
Length=183
Score = 39.7 bits (91), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 42/102 (41%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 80 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 129
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+I + + ND+ W D +F+ VLE
Sbjct 130 KELLPFIRTQVVGDFTATRVNDT----AWVDGKFVVTEEVLE 167
> stl:stu0066 hypothetical protein
Length=183
Score = 39.7 bits (91), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 42/102 (41%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 80 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 129
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+I + + ND+ W D +F+ VLE
Sbjct 130 KELLPFIRTQVVGDFTATRVNDT----AWVDGKFVVTEEVLE 167
> stc:str0066 hypothetical protein
Length=183
Score = 39.7 bits (91), Expect = 0.24, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 42/102 (41%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 80 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 129
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+I + + ND+ W D +F+ VLE
Sbjct 130 KELLPFIRTQVVGDFTATRVNDT----AWVDGKFVVTEEVLE 167
> stj:SALIVA_0090 flavoprotein
Length=181
Score = 39.3 bits (90), Expect = 0.30, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 43/102 (42%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 80 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 129
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+ +I + + ND+ W D +F+ VLE
Sbjct 130 KDLLPFIRTQVVGDFTATRVNDT----AWADGKFLATEEVLE 167
> ssr:SALIVB_0097 Oxidoreductase (EC:1.1.1.-)
Length=181
Score = 39.3 bits (90), Expect = 0.36, Method: Compositional matrix adjust.
Identities = 27/102 (26%), Positives = 43/102 (42%), Gaps = 14/102 (14%)
Query 116 TIPSPIADVLLNLQSTFDLSEHCLHPEDFSALHDVLYASKRFCWLCSWLNMSHKQYFEIW 175
+IP P+ ++L L DLS+ P SAL D + + S N H Q F+ +
Sbjct 80 SIPGPVKNLLDWLSRALDLSD----PSGPSALQD------KIVTVSSVANGGHNQLFDAY 129
Query 176 SNFYKYINLSLYKEHYLALENDSKYFRGWFDRRFITPTAVLE 217
+ +I + + ND+ W D +F+ VLE
Sbjct 130 KDLLPFIRTQVVGDFTATRVNDT----AWADGKFLATEEVLE 167
> crb:CARUB_v10016366mg hypothetical protein
Length=1135
Score = 37.0 bits (84), Expect = 4.8, Method: Composition-based stats.
Identities = 23/78 (29%), Positives = 35/78 (45%), Gaps = 15/78 (19%)
Query 96 SNQVQW-LKNWYYYYY------------SRPLETIPSPIADVLLNLQSTFDLSE--HCLH 140
SNQ+ W + W +Y+ S PL+T P P++ VL N+ F E H +
Sbjct 809 SNQLPWDCQRWNMFYFLWGVFRGKKKSCSNPLKTTPLPVSCVLPNMGKAFSSRETFHHEN 868
Query 141 PEDFSALHDVLYASKRFC 158
P D +L D + + C
Sbjct 869 PSDRESLTDCTSSRMQLC 886
> ath:AT5G63750 ARI13; putative E3 ubiquitin-protein ligase ARI13
Length=536
Score = 36.6 bits (83), Expect = 5.4, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 32/66 (48%), Gaps = 8/66 (12%)
Query 125 LLNLQSTFDLSEHCLHPEDFSALHDVLY----ASKRFCWLCSWLNMSHKQYFEIWSNFYK 180
L ++++T + HCL P D + + + RFCW C +HK S FYK
Sbjct 267 LSSIKATKKVCPHCLRPADLGTKQYLRFLTCACNGRFCWKCMQPEEAHKTE----SGFYK 322
Query 181 YINLSL 186
+ N+S+
Sbjct 323 FCNVSM 328
> dmu:Desmu_1040 enolase
Length=435
Score = 35.8 bits (81), Expect = 8.6, Method: Compositional matrix adjust.
Identities = 16/53 (30%), Positives = 29/53 (55%), Gaps = 3/53 (6%)
Query 105 WYYYYYSRPLETIPSPIADVL---LNLQSTFDLSEHCLHPEDFSALHDVLYAS 154
+YYY R +T+P P+ +++ ++ + D E + P FS+ HD L A+
Sbjct 137 FYYYLGGRAADTLPVPLLNIINGGVHAGNKLDFQEFMIVPAGFSSFHDALKAA 189
Lambda K H a alpha
0.325 0.138 0.449 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 375861744714