bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-6_CDS_annotation_glimmer3.pl_2_3
Length=61
Score E
Sequences producing significant alignments: (Bits) Value
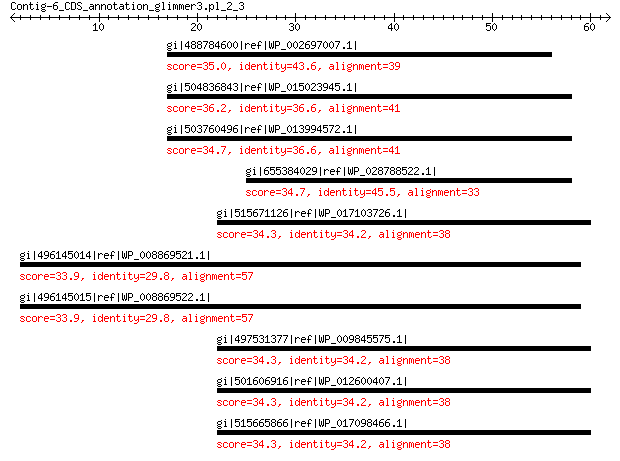
gi|488784600|ref|WP_002697007.1| hypothetical protein 35.0 0.42
gi|504836843|ref|WP_015023945.1| nuclease 36.2 0.56
gi|503760496|ref|WP_013994572.1| nuclease 34.7 2.1
gi|655384029|ref|WP_028788522.1| nuclease 34.7 2.2
gi|515671126|ref|WP_017103726.1| membrane protein 34.3 4.8
gi|496145014|ref|WP_008869521.1| hypothetical protein 33.9 4.9
gi|496145015|ref|WP_008869522.1| hypothetical protein 33.9 5.1
gi|497531377|ref|WP_009845575.1| membrane protein 34.3 5.2
gi|501606916|ref|WP_012600407.1| membrane protein 34.3 5.2
gi|515665866|ref|WP_017098466.1| membrane protein 34.3 5.7
>gi|488784600|ref|WP_002697007.1| hypothetical protein [Microscilla marina]
gi|123989328|gb|EAY28889.1| conserved hypothetical protein [Microscilla marina ATCC 23134]
Length=91
Score = 35.0 bits (79), Expect = 0.42, Method: Compositional matrix adjust.
Identities = 17/39 (44%), Positives = 24/39 (62%), Gaps = 0/39 (0%)
Query 17 NVKTGAIDYLMYFDWSARAINEFITDFVDSNKDYVVRAF 55
N+K DYL F A++ ++FITD +D N D V+AF
Sbjct 16 NIKDFPNDYLATFGLVAKSADDFITDTIDLNPDKAVQAF 54
>gi|504836843|ref|WP_015023945.1| nuclease [Psychroflexus torquis]
gi|408490722|ref|YP_006867091.1| putative nuclease, PIN_SF superfamily protein [Psychroflexus
torquis ATCC 700755]
gi|408467997|gb|AFU68341.1| putative nuclease, PIN_SF superfamily protein [Psychroflexus
torquis ATCC 700755]
Length=187
Score = 36.2 bits (82), Expect = 0.56, Method: Compositional matrix adjust.
Identities = 15/41 (37%), Positives = 26/41 (63%), Gaps = 0/41 (0%)
Query 17 NVKTGAIDYLMYFDWSARAINEFITDFVDSNKDYVVRAFKQ 57
N+K DYL F +A+ ++F+TD +D N + V+AF++
Sbjct 112 NIKHFPKDYLASFGLTAKTADDFLTDIIDLNPEQAVKAFRE 152
>gi|503760496|ref|WP_013994572.1| nuclease [Zobellia galactanivorans]
gi|340619204|ref|YP_004737657.1| hypothetical protein zobellia_3240 [Zobellia galactanivorans]
gi|339734001|emb|CAZ97378.1| Conserved hypothetical protein [Zobellia galactanivorans]
Length=187
Score = 34.7 bits (78), Expect = 2.1, Method: Compositional matrix adjust.
Identities = 15/41 (37%), Positives = 25/41 (61%), Gaps = 0/41 (0%)
Query 17 NVKTGAIDYLMYFDWSARAINEFITDFVDSNKDYVVRAFKQ 57
N+K YL F SA+ ++F+TD +D N + ++AFK+
Sbjct 112 NIKDFPKAYLSSFGLSAKTADDFLTDIIDLNPEEAIKAFKE 152
>gi|655384029|ref|WP_028788522.1| nuclease [Terrimonas ferruginea]
Length=187
Score = 34.7 bits (78), Expect = 2.2, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 23/33 (70%), Gaps = 0/33 (0%)
Query 25 YLMYFDWSARAINEFITDFVDSNKDYVVRAFKQ 57
YL F SA++ ++F+TD +D N+D V AFK+
Sbjct 120 YLQSFGLSAKSADDFLTDIIDINQDQAVDAFKE 152
>gi|515671126|ref|WP_017103726.1| membrane protein [Vibrio tasmaniensis]
Length=728
Score = 34.3 bits (77), Expect = 4.8, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 26/38 (68%), Gaps = 1/38 (3%)
Query 22 AIDYLMYFDWSARAINEFITDFVDSNKDYVVRAFKQYK 59
A+ Y++ DW ++ +++ + D +DSNKDY+ + QY+
Sbjct 525 AVSYILP-DWQSKRLHKVMADALDSNKDYLAQIIGQYR 561
>gi|496145014|ref|WP_008869521.1| hypothetical protein [Desulfonatronospira thiodismutans]
gi|298510288|gb|EFI34191.1| protein of unknown function DUF1568 [Desulfonatronospira thiodismutans
ASO3-1]
Length=306
Score = 33.9 bits (76), Expect = 4.9, Method: Compositional matrix adjust.
Identities = 17/57 (30%), Positives = 28/57 (49%), Gaps = 7/57 (12%)
Query 2 IMKTERKFTWCVRFYNVKTGAIDYLMYFDWSARAINEFITDFVDSNKDYVVRAFKQY 58
I+K + WC Y+ +TG D L+ D+ + NEF N +VR ++Q+
Sbjct 165 IVKKPEDYRWCSLGYHTQTGNKDGLLSIDFGMKEWNEF-------NPSEIVRKYRQF 214
>gi|496145015|ref|WP_008869522.1| hypothetical protein [Desulfonatronospira thiodismutans]
gi|298510289|gb|EFI34192.1| protein of unknown function DUF1568 [Desulfonatronospira thiodismutans
ASO3-1]
gi|298510292|gb|EFI34195.1| protein of unknown function DUF1568 [Desulfonatronospira thiodismutans
ASO3-1]
Length=304
Score = 33.9 bits (76), Expect = 5.1, Method: Compositional matrix adjust.
Identities = 17/57 (30%), Positives = 28/57 (49%), Gaps = 7/57 (12%)
Query 2 IMKTERKFTWCVRFYNVKTGAIDYLMYFDWSARAINEFITDFVDSNKDYVVRAFKQY 58
I+K + WC Y+ +TG D L+ D+ + NEF N +VR ++Q+
Sbjct 165 IVKKPEDYRWCSLGYHTQTGNKDGLLSIDFGMKEWNEF-------NPSEIVRKYRQF 214
>gi|497531377|ref|WP_009845575.1| membrane protein [Vibrio sp. MED222]
gi|85837228|gb|EAQ55340.1| Predicted membrane protein [Vibrio sp. MED222]
Length=728
Score = 34.3 bits (77), Expect = 5.2, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 26/38 (68%), Gaps = 1/38 (3%)
Query 22 AIDYLMYFDWSARAINEFITDFVDSNKDYVVRAFKQYK 59
A+ Y++ DW ++ +++ + D +DSNKDY+ + QY+
Sbjct 525 AVSYILP-DWQSKRLHKVMADALDSNKDYLAQIIGQYR 561
>gi|501606916|ref|WP_012600407.1| membrane protein [Vibrio splendidus]
gi|218676238|ref|YP_002395057.1| membrane protein [Vibrio splendidus LGP32]
gi|218324506|emb|CAV25973.1| Predicted membrane protein [Vibrio splendidus LGP32]
Length=728
Score = 34.3 bits (77), Expect = 5.2, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 26/38 (68%), Gaps = 1/38 (3%)
Query 22 AIDYLMYFDWSARAINEFITDFVDSNKDYVVRAFKQYK 59
A+ Y++ DW ++ +++ + D +DSNKDY+ + QY+
Sbjct 525 AVSYILP-DWQSKRLHKVMADALDSNKDYLAQIIGQYR 561
>gi|515665866|ref|WP_017098466.1| membrane protein [Vibrio tasmaniensis]
Length=728
Score = 34.3 bits (77), Expect = 5.7, Method: Composition-based stats.
Identities = 13/38 (34%), Positives = 26/38 (68%), Gaps = 1/38 (3%)
Query 22 AIDYLMYFDWSARAINEFITDFVDSNKDYVVRAFKQYK 59
A+ Y++ DW ++ +++ + D +DSNKDY+ + QY+
Sbjct 525 AVSYILP-DWQSKRLHKVMADALDSNKDYLAQIIGQYR 561
Lambda K H a alpha
0.331 0.140 0.453 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 429216838839