bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-5_CDS_annotation_glimmer3.pl_2_2
Length=84
Score E
Sequences producing significant alignments: (Bits) Value
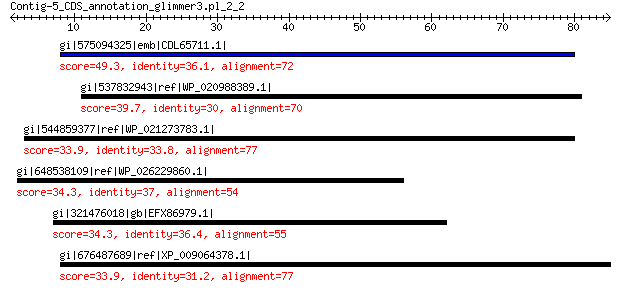
gi|575094325|emb|CDL65711.1| unnamed protein product 49.3 9e-06
gi|537832943|ref|WP_020988389.1| putative large structural protein 39.7 0.16
gi|544859377|ref|WP_021273783.1| hypothetical protein 33.9 2.3
gi|648538109|ref|WP_026229860.1| endopolygalacturonase 34.3 6.3
gi|321476018|gb|EFX86979.1| hypothetical protein DAPPUDRAFT_312451 34.3 6.6
gi|676487689|ref|XP_009064378.1| hypothetical protein LOTGIDRAFT... 33.9 7.9
>gi|575094325|emb|CDL65711.1| unnamed protein product [uncultured bacterium]
Length=107
Score = 49.3 bits (116), Expect = 9e-06, Method: Compositional matrix adjust.
Identities = 26/72 (36%), Positives = 44/72 (61%), Gaps = 5/72 (7%)
Query 8 LAITPSQVQELTNKGISVSLPNAKQFLDGNSLETAKSWNVDPIFRRDANICTMFELERDS 67
LAITPS +++L +G+ VS+PNA F +S V P + DA+ T++EL + S
Sbjct 37 LAITPSDIEKLARQGVPVSVPNANSFYSIDS-----GLEVPPELKVDADRNTLWELSQQS 91
Query 68 QGKIIRAHKVDR 79
+ +I++A K ++
Sbjct 92 KARIMKARKREK 103
>gi|537832943|ref|WP_020988389.1| putative large structural protein [Leptospira inadai]
gi|529313970|gb|EQA37700.1| putative large structural protein [Leptospira inadai serovar
Lyme str. 10]
Length=2768
Score = 39.7 bits (91), Expect = 0.16, Method: Composition-based stats.
Identities = 21/70 (30%), Positives = 37/70 (53%), Gaps = 0/70 (0%)
Query 11 TPSQVQELTNKGISVSLPNAKQFLDGNSLETAKSWNVDPIFRRDANICTMFELERDSQGK 70
T +++QELT+K + + + L + WN D I RDAN+ F ++D GK
Sbjct 2534 TQTKIQELTSKYENGEINKGIYESEKTKLNQIRDWNSDVIALRDANLVKEFVFQKDKSGK 2593
Query 71 IIRAHKVDRK 80
+I++ ++ K
Sbjct 2594 LIQSRELALK 2603
>gi|544859377|ref|WP_021273783.1| hypothetical protein [Bacteriovorax sp. Seq25_V]
gi|530767439|gb|EQC46520.1| hypothetical protein M900_2504 [Bacteriovorax sp. Seq25_V]
Length=93
Score = 33.9 bits (76), Expect = 2.3, Method: Compositional matrix adjust.
Identities = 26/83 (31%), Positives = 36/83 (43%), Gaps = 11/83 (13%)
Query 3 VTKPDLAITPSQVQELTNKGISVSLPNA--KQFLDGNSLETA----KSWNVDPIFRRDAN 56
KP+ A+ S + N + LP + +QF+ G T K DPIF + N
Sbjct 11 CVKPN-ALVQSDDHSVYNSVVRTYLPQSTHQQFIGGRGCSTGQGELKRLRFDPIFTLNHN 69
Query 57 ICTMFELERDSQGKIIRAHKVDR 79
F + RD GK+IR H R
Sbjct 70 ----FAMLRDGIGKLIRRHGGRR 88
>gi|648538109|ref|WP_026229860.1| endopolygalacturonase [Burkholderia sp. JPY347]
Length=715
Score = 34.3 bits (77), Expect = 6.3, Method: Composition-based stats.
Identities = 20/58 (34%), Positives = 28/58 (48%), Gaps = 4/58 (7%)
Query 2 LVTKPDLAITPSQVQELTNKGISVSLPNAKQFL----DGNSLETAKSWNVDPIFRRDA 55
L P+ + PS V LT G+ V PN F +GN L T +++N D + DA
Sbjct 369 LSNSPNFHVVPSGVDGLTIWGVKVQTPNLAAFANPAGNGNPLYTGQTFNRDNVKNTDA 426
>gi|321476018|gb|EFX86979.1| hypothetical protein DAPPUDRAFT_312451 [Daphnia pulex]
Length=432
Score = 34.3 bits (77), Expect = 6.6, Method: Compositional matrix adjust.
Identities = 20/58 (34%), Positives = 33/58 (57%), Gaps = 10/58 (17%)
Query 7 DLAITPSQVQELTNKGI---SVSLPNAKQFLDGNSLETAKSWNVDPIFRRDANICTMF 61
D+ ++P+ V+ L + G+ + L A FL+GNS N+DPI R ++N T+F
Sbjct 83 DIMLSPNPVKSLLDNGMDKATTFLIGANNFLNGNS-------NIDPIDRFNSNFNTLF 133
>gi|676487689|ref|XP_009064378.1| hypothetical protein LOTGIDRAFT_168254 [Lottia gigantea]
gi|556096341|gb|ESO84993.1| hypothetical protein LOTGIDRAFT_168254 [Lottia gigantea]
Length=416
Score = 33.9 bits (76), Expect = 7.9, Method: Composition-based stats.
Identities = 24/78 (31%), Positives = 41/78 (53%), Gaps = 12/78 (15%)
Query 8 LAITPSQVQELTNKGISVSLPNAKQFLDGNSLETAKSWNVDPIFRRDANICTMFELERD- 66
+T ++E + + I + + +FL N+LE VD IFRR A T+ +++RD
Sbjct 227 FGVTLEYIKENSGRIIPIVVEQTVEFLKENALE------VDGIFRRSARAVTLKQVQRDF 280
Query 67 SQGKIIRAHKVDRKKFGD 84
++GK +VD + GD
Sbjct 281 NEGK-----EVDFNEIGD 293
Lambda K H a alpha
0.317 0.133 0.383 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 431162850036