bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_3
Length=569
Score E
Sequences producing significant alignments: (Bits) Value
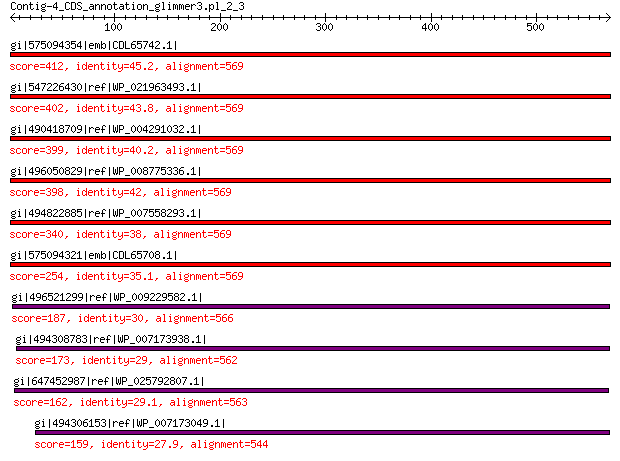
gi|575094354|emb|CDL65742.1| unnamed protein product 412 1e-132
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 402 3e-129
gi|490418709|ref|WP_004291032.1| hypothetical protein 399 7e-128
gi|496050829|ref|WP_008775336.1| hypothetical protein 398 9e-128
gi|494822885|ref|WP_007558293.1| hypothetical protein 340 8e-105
gi|575094321|emb|CDL65708.1| unnamed protein product 254 4e-72
gi|496521299|ref|WP_009229582.1| capsid protein 187 1e-48
gi|494308783|ref|WP_007173938.1| hypothetical protein 173 1e-43
gi|647452987|ref|WP_025792807.1| hypothetical protein 162 9e-40
gi|494306153|ref|WP_007173049.1| hypothetical protein 159 5e-39
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 412 bits (1059), Expect = 1e-132, Method: Compositional matrix adjust.
Identities = 257/631 (41%), Positives = 354/631 (56%), Gaps = 83/631 (13%)
Query 1 VKNHPRRSGFDLSSKVAFTAKVGELLPVKWCLTMPGDKFSLSEQHFTRTQPVNTSAYTRV 60
+KN P R+GFDLS K FTAK GELLPV + +PGD F+++ + FTRTQP+NTSA+ R+
Sbjct 6 IKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTSAFARM 65
Query 61 REYYDWFWCPLHLLWRNAPEVIAQIQQNVQHAS--SYDGSVLLGSNMPCVslsqls--kl 116
REYYD+++ P +W I Q+ NVQHAS + D + L MP + Q++
Sbjct 66 REYYDFYFVPFEQMWNKFDSCITQMNANVQHASGPTLDDNTPLSGRMPYFTSEQIADYLN 125
Query 117 lsslkgkkNYFGFDRSDLAYKILQYLRYGNVQTSSSTSGKNFGTSIPLSDRSYSQDYVFN 176
+ +KN FGF+RS L K+LQYL YG+ + S + N ++ PL ++N
Sbjct 126 DQATAARKNPFGFNRSTLTCKLLQYLGYGDYNSFDSET--NTWSAKPL---------LYN 174
Query 177 HALSIFPLLGYKKFCQDYFRFTQWQDSAPYLWNIDYYDAKKSTSILPDTFSTSYLTHNTL 236
LS FPLL Y+K D++R+TQW+ + P +N+DY K TS L + N
Sbjct 175 LELSPFPLLAYQKIYSDFYRYTQWEKTNPSTFNLDYI---KGTSDLQMDLTGLPSDDNNF 231
Query 237 IDMEYCNWNKDMFFGVLPDAQYGDASVVDI------------------------------ 266
D+ YCN+ KDMF GVLP AQYG ASVV I
Sbjct 232 FDIRYCNYQKDMFHGVLPVAQYGSASVVPINGQLNVISNGDSGPIFKTSTPDPGTPGTSY 291
Query 267 ------------SFGMSGQTVVASPSDISSRYTISNPSDSST-------PNL---SGSPL 304
SFG+SG T+ S S Y PS++ST PNL +
Sbjct 292 VTVGGNIGVDNRSFGVSGSTLNVGKSADPSGYGF--PSNASTRSLLWENPNLIIENNQGF 349
Query 305 VLDVLALRRGEALQRFREISLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDIS 364
+ +LALR+ E LQ+++E+S+ +Y+SQI+ H+G+ V LS + Y+GG A+SLDI+
Sbjct 350 YVPILALRQAEFLQKWKEVSVSGEEDYKSQIEKHWGIKVSDFLSHQARYLGGCATSLDIN 409
Query 365 EVVNTNITESNEALIAGKGIGTGQFSDKFYAK-DWGILMCIYHSVPLLDYVLTSPDPQLF 423
EV+N NIT N A IAGKG TG S +F +K ++GI+MCIYH +P++DYV + D
Sbjct 410 EVINNNITGDNAADIAGKGTFTGNGSIRFESKGEYGIIMCIYHVLPIVDYVGSGVDHSCT 469
Query 424 LSENTSFPVPELDAIGLESIPLSCYSNSSLEIPITNPNVDAASLTMGYLPRYYAWKTSLD 483
L + TSFP+PELD IG+ES+PL N P+ + +A +GY PRY WKTS+D
Sbjct 470 LVDATSFPIPELDQIGMESVPLVRAMN-----PVKESDTPSADTFLGYAPRYIDWKTSVD 524
Query 484 YVLGAFTTTEKEWVAPI-TASLWSKMLLPV----TVDGSGINYNFFKVNPSILDPIFLVN 538
+G F + + W P+ L S L V+ I FFKVNPSI+DP+F V
Sbjct 525 RSVGDFADSLRTWCLPVGDKELTSANSLNFPSNPNVEPDSIAAGFFKVNPSIVDPLFAVV 584
Query 539 ADSTWDTDTFLVNAAFDIRVARNLDYDGMPY 569
ADST TD FL ++ FD++V RNLD +G+PY
Sbjct 585 ADSTVKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 402 bits (1032), Expect = 3e-129, Method: Compositional matrix adjust.
Identities = 249/593 (42%), Positives = 341/593 (58%), Gaps = 53/593 (9%)
Query 1 VKNHPRRSGFDLSSKVAFTAKVGELLPVKWCLTMPGDKFSLSEQHFTRTQPVNTSAYTRV 60
+KN +R+GFDLS K AFTAKVGELLP+ PGDKF++ Q FTRTQPVN++AY+R+
Sbjct 10 LKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQPVNSAAYSRL 69
Query 61 REYYDWFWCPLHLLWRNAPEVIAQIQQNVQHASSYDGSVLLGSNMPCVs--------lsq 112
REYYD+++ P LLW AP + + HA+ SV L P + +
Sbjct 70 REYYDFYFVPYRLLWNMAPTFFTNM-PDPHHAADLVSSVNLSQRHPWFTFFDIMEYLGNL 128
Query 113 lskllsslkgkkNYFGFDRSDLAYKILQYLRYGNVQTSSSTSGKNFGTSIPLSDRSYSQD 172
S + K +KN+FGF R +L+ K+L YL YG GK++ + SD S D
Sbjct 129 NSLSGAYEKYQKNFFGFSRVELSVKLLNYLNYG--------FGKDYESVKVPSD---SDD 177
Query 173 YVFNHALSIFPLLGYKKFCQDYFRFTQWQDSAPYLWNIDYYDAKKSTSILP-DTFSTSYL 231
V LS FPLL Y+K C+DYFR QWQ +APY +N+DY K S +P +F+
Sbjct 178 IV----LSPFPLLAYQKICEDYFRDDQWQSAAPYRYNLDYLYGKSSGFHIPMSSFTNDAF 233
Query 232 THNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDISFG------MSGQTVVASPSD---- 281
+ T+ D+ YCN+ KD F G+LP AQYGD SV FG S T ++P
Sbjct 234 KNPTMFDLNYCNFQKDYFTGMLPRAQYGDVSVASPIFGDLDIGDSSSLTFASAPQQGANT 293
Query 282 ISSRYTISNPSDSSTPNLSGSPLVLDVLALRRGEALQRFREISLCTPANYRSQIKAHFGV 341
I S + N + ++T LS VLALR+ E LQ++REI+ +Y++Q++ HF V
Sbjct 294 IQSGVLVVNNNSNTTAGLS-------VLALRQAECLQKWREIAQSGKMDYQTQMQKHFNV 346
Query 342 DVGSELSGMSTYIGGEASSLDISEVVNTNITESNEALIAGKGIGT--GQFSDKFYAKDWG 399
+ LSG Y+GG S+LDISEVVNTN+T N+A I GKG GT G D F + + G
Sbjct 347 SPSATLSGHCKYLGGWTSNLDISEVVNTNLTGDNQADIQGKGTGTLNGNKVD-FESSEHG 405
Query 400 ILMCIYHSVPLLDYVLTSPDPQLFLSENTSFPVPELDAIGLESIPLSCYSNSSLEIPITN 459
I+MCIYH +PLLD+ + Q F + T + +PE D++G++ + S ++P
Sbjct 406 IIMCIYHCLPLLDWSINRIARQNFKTTFTDYAIPEFDSVGMQQLYPSEMIFGLEDLP--- 462
Query 460 PNVDAASLTMGYLPRYYAWKTSLDYVLGAFTTTEKEWVAPITASLWSKMLLPVTVDGSG- 518
D +S+ MGY+PRY KTS+D + G+F T WV+P+T S S G
Sbjct 463 --SDPSSINMGYVPRYADLKTSIDEIHGSFIDTLVSWVSPLTDSYISAYRQACKDAGFSD 520
Query 519 --INYNFFKVNPSILDPIFLVNADSTWDTDTFLVNAAFDIRVARNLDYDGMPY 569
+ YNFFKVNP I+D IF V ADST +TD L+N+ FDI+ RN DY+G+PY
Sbjct 521 ITMTYNFFKVNPHIVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYNGLPY 573
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 399 bits (1024), Expect = 7e-128, Method: Compositional matrix adjust.
Identities = 229/595 (38%), Positives = 340/595 (57%), Gaps = 52/595 (9%)
Query 1 VKNHPRRSGFDLSSKVAFTAKVGELLPVKWCLTMPGDKFSLSEQHFTRTQPVNTSAYTRV 60
++N P R+GFDLS K FTAK GELLPV +PGD F ++ + FTRTQPVNT+A+ R+
Sbjct 10 IRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQPVNTAAFARI 69
Query 61 REYYDWFWCPLHLLWRNAPEVIAQIQQNVQHASSYDGS--VLLGSNMPCVslsqls---- 114
REYYD+F+ P LLW A V+ Q+ N QHA S D + +L MP ++ ++
Sbjct 70 REYYDFFFVPYDLLWNKANTVLTQMYDNPQHAVSIDPTRNFVLSGEMPYMTSEAIASYIN 129
Query 115 ---kllsslkgkkNYFGFDRSDLAYKILQYLRYGNVQTSSSTSGKNFGTSIPLSDRSYSQ 171
+ K NYFG++RS + K+L+YL YGN ++ L+D +
Sbjct 130 ALSTASALADYKSNYFGYNRSKSSVKLLEYLGYGNYESF-------------LTDDWNTA 176
Query 172 DYVFNHALSIFPLLGYKKFCQDYFRFTQWQDSAPYLWNIDYYDAKKSTSILPDTFSTSYL 231
+ N +IF LL Y+K D++R +QW+ +P +N+DY D S+ L + +ST +
Sbjct 177 PLMANLNHNIFGLLAYQKIYSDFYRDSQWERVSPSTFNVDYLDG--SSMNLDNAYSTEFY 234
Query 232 THNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDISFGMSGQTVVASPSDISSRYTISNP 291
+ D+ YCNW KD+F GVLP QYG+ +V I+ ++G+ +++ S + + T +
Sbjct 235 QNYNFFDLRYCNWQKDLFHGVLPHQQYGETAVASITPDVTGKLTLSNFSTVGTSPTTA-- 292
Query 292 SDSSTPNLSGSPLV--LDVLALRRGEALQRFREISLCTPANYRSQIKAHFGVDVGSELSG 349
S ++T NL V L +L LR+ E LQ+++EI+ +Y+ Q++ H+GV VG S
Sbjct 293 SGTATKNLPAFDTVGDLSILVLRQAEFLQKWKEITQSGNKDYKDQLEKHWGVSVGDGFSE 352
Query 350 MSTYIGGEASSLDISEVVNTNITESNEALIAGKGIGTGQFSDKFYAKD-WGILMCIYHSV 408
+ TY+GG +SS+DI+EV+NTNIT S A IAGKG+G F + +G++MCIYH +
Sbjct 353 LCTYLGGVSSSIDINEVINTNITGSAAADIAGKGVGVANGEINFNSNGRYGLIMCIYHCL 412
Query 409 PLLDYVLTSPDPQLFLSENTSFPVPELDAIGLESIPLSCYSNSSLEIPITNP---NVDAA 465
PLLDY DP +T + +PE D +G++S+PL + + NP +A+
Sbjct 413 PLLDYTTDMLDPAFLKVNSTDYAIPEFDRVGMQSMPL---------VQLMNPLRSFANAS 463
Query 466 SLTMGYLPRYYAWKTSLDYVLGAFTTTEKEWVAPI-TASLWSKMLLPVTV---------- 514
L +GY+PRY +KTS+D +G F T WV S+ ++ LP
Sbjct 464 GLVLGYVPRYIDYKTSVDQSVGGFKRTLNSWVISYGNISVLKQVTLPNDAPPIEPSEPVP 523
Query 515 DGSGINYNFFKVNPSILDPIFLVNADSTWDTDTFLVNAAFDIRVARNLDYDGMPY 569
+ +N+ FFKVNP LDPIF V A +TD FL ++ FDI+ RNLD DG+PY
Sbjct 524 SVAPMNFTFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLDTDGLPY 578
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 398 bits (1023), Expect = 9e-128, Method: Compositional matrix adjust.
Identities = 239/593 (40%), Positives = 346/593 (58%), Gaps = 46/593 (8%)
Query 1 VKNHPRRSGFDLSSKVAFTAKVGELLPVKWCLTMPGDKFSLSEQHFTRTQPVNTSAYTRV 60
++N R+GFDLSSK FTAK GELLPVK +PGDK+S+ + FTRTQP+NT+A+ R+
Sbjct 10 LRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEVLPGDKWSIDLKSFTRTQPLNTAAFARM 69
Query 61 REYYDWFWCPLHLLWRNAPEVIAQIQQNVQHASSY--DGSVLLGSNMPCVslsqls---- 114
REYYD+++ P +LLW A V+ Q+ N QHA+SY + L MP V+ ++
Sbjct 70 REYYDFYFVPYNLLWNKANTVLTQMYDNPQHATSYIPSANQALAGVMPNVTCKGIADYLN 129
Query 115 ----kllsslkgkkNYFGFDRSDLAYKILQYLRYGNVQTSSSTSGKNFGTSIPLSDRSYS 170
+ ++ +KNYFG+ RS K+L+YL YGN T +TS N T PLS
Sbjct 130 LVAPDVTTTNSYEKNYFGYSRSLGTAKLLEYLGYGNFYT-YATSKNNTWTKSPLSS---- 184
Query 171 QDYVFNHALSIFPLLGYKKFCQDYFRFTQWQDSAPYLWNIDYYDAKKSTSILPDTFST-- 228
N L+I+ +L Y+K D+ R +QW+ +P +N+DY +++ D+ T
Sbjct 185 -----NLQLNIYGVLAYQKIYADHIRDSQWEKVSPSCFNVDYLSGTVDSAMTIDSMITGQ 239
Query 229 SYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDISFG--MSGQTVVASPSD--ISS 284
+ + D+ YCNW KD+F GVLP QYGD + V+++ +S Q +V +P +
Sbjct 240 GFAPFYNMFDLRYCNWQKDLFHGVLPRQQYGDTAAVNVNLSNVLSAQYMVQTPDGDPVGG 299
Query 285 RYTISNPSDSSTPNLSGSPLVLDVLALRRGEALQRFREISLCTPANYRSQIKAHFGVDVG 344
S + T N SG+ VLALR+ E LQ+++EI+ +Y+ QI+ H+ V VG
Sbjct 300 SPFSSTGVNLQTVNGSGT---FTVLALRQAEFLQKWKEITQSGNKDYKDQIEKHWNVSVG 356
Query 345 SELSGMSTYIGGEASSLDISEVVNTNITESNEALIAGKGIGTGQFSDKFYAKD-WGILMC 403
S MS Y+GG +SLDI+EVVN NIT SN A IAGKG+ G F A + +G++MC
Sbjct 357 EAYSEMSLYLGGTTASLDINEVVNNNITGSNAADIAGKGVVVGNGRISFDAGERYGLIMC 416
Query 404 IYHSVPLLDYVLTSPDPQLFLSENTSFPVPELDAIGLESIPLSCYSNSSLEIPITNP--- 460
IYHS+PLLDY +P +T F +PE D +G+ES+PL + + NP
Sbjct 417 IYHSLPLLDYTTDLVNPAFTKINSTDFAIPEFDRVGMESVPL---------VSLMNPLQS 467
Query 461 NVDAASLTMGYLPRYYAWKTSLDYVLGAFTTTEKEWVAPI-TASLWSKMLL---PVTVDG 516
+ + S +GY PRY ++KT +D +GAF TT K WV S+ +++ P G
Sbjct 468 SYNVGSSILGYAPRYISYKTDVDSSVGAFKTTLKSWVMSYDNQSVINQLNYQDDPNNSPG 527
Query 517 SGINYNFFKVNPSILDPIFLVNADSTWDTDTFLVNAAFDIRVARNLDYDGMPY 569
+ +NY FKVNP+ +DP+F V A ++ DTD FL ++ FD++V RNLD DG+PY
Sbjct 528 TLVNYTNFKVNPNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 340 bits (871), Expect = 8e-105, Method: Compositional matrix adjust.
Identities = 216/617 (35%), Positives = 320/617 (52%), Gaps = 68/617 (11%)
Query 1 VKNHPRRSGFDLSSKVAFTAKVGELLPVKWCLTMPGDKFSLSEQHFTRTQPVNTSAYTRV 60
V+N P R+G+DL+ K+ FTAK G L+PV W +P D + + + F RTQP+NT+A+ R+
Sbjct 17 VRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQPLNTAAFARM 76
Query 61 REYYDWFWCPLHLLWRNAPEVIAQIQQNVQHASS--YDGSVLLGSNMPCVslsqlsklls 118
R Y+D+++ P +W P I Q++ N+ HAS +V L +P + Q++ +
Sbjct 77 RGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLADNVPLSDELPYFTAEQVADYIV 136
Query 119 slkgkkNYFGFDRSDLAYKILQYLRYGN----VQTSSSTSGKNFGTSIPLSDRSYSQDYV 174
SL KN FG+ R+ L IL+YL YG+ + ++ G + T L+
Sbjct 137 SLADSKNQFGYYRAWLVCIILEYLGYGDFYPYIVEAAGGEGATWATRPMLN--------- 187
Query 175 FNHALSIFPLLGYKKFCQDYFRFTQWQDSAPYLWNIDYYDAKKSTSILPDTFSTSYLTHN 234
N S FPL Y+K D+ R+TQW+ S P +NIDY + + S+ D +
Sbjct 188 -NLKFSPFPLFAYQKIYADFNRYTQWERSNPSTFNIDYI-SGSADSLQLDFTVEGFKDSF 245
Query 235 TLIDMEYCNWNKDMFFGVLPDAQYGDASVVDISFGM------------SGQTVV------ 276
L DM Y NW +D+ G +P AQYG+AS V +S M +GQ V
Sbjct 246 NLFDMRYSNWQRDLLHGTIPQAQYGEASAVPVSGSMQVVEGPTPPAFTTGQDGVAFLNGN 305
Query 277 -----------ASPSDISSRYTISNPSDSSTPNLSGSPLVLDVLALRRGEALQRFREISL 325
A S SR N ++S S + +LALRR EA Q+++E++L
Sbjct 306 VTIQGSSGYLQAQTSVGESRILRFNNTNSGLIVEGDSSFGVSILALRRAEAAQKWKEVAL 365
Query 326 CTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNITESNEALIAGKGIG 385
+ +Y SQI+AH+G V S M ++G L I+EVVN NIT N A IAGKG
Sbjct 366 ASEEDYPSQIEAHWGQSVNKAYSDMCQWLGSINIDLSINEVVNNNITGENAADIAGKGTM 425
Query 386 TGQFSDKF-YAKDWGILMCIYHSVPLLDYVLTSPDPQLFLSENTSFPVPELDAIGLESIP 444
+G S F +GI+MC++H +P LDY+ ++P L+ FP+PE D IG+E +P
Sbjct 426 SGNGSINFNVGGQYGIVMCVFHVLPQLDYITSAPHFGTTLTNVLDFPIPEFDKIGMEQVP 485
Query 445 LSCYSNSSLEIPITNPNVD---AASLTMGYLPRYYAWKTSLDYVLGAFTTTEKEWVAPIT 501
+ N P+ + D + +L GY P+YY WKT+LD +G F + K W+ P
Sbjct 486 VIRGLN-----PVKPKDGDFKVSPNLYFGYAPQYYNWKTTLDKSMGEFRRSLKTWIIPFD 540
Query 502 ASLWSKMLLPV---------TVDGSGINYNFFKVNPSILDPIFLVNADSTWDTDTFLVNA 552
+ LL V+ + FFKV+PS+LD +F V A+S +TD FL +
Sbjct 541 ----DEALLAADSVDFPDNPNVEADSVKAGFFKVSPSVLDNLFAVKANSDLNTDQFLCST 596
Query 553 AFDIRVARNLDYDGMPY 569
FD+ V R+LD +G+PY
Sbjct 597 LFDVNVVRSLDPNGLPY 613
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 254 bits (648), Expect = 4e-72, Method: Compositional matrix adjust.
Identities = 200/648 (31%), Positives = 301/648 (46%), Gaps = 99/648 (15%)
Query 1 VKNHPRRSGFDLSSKVAFTAKVGELLPVKWCLTMPGDKFSLSEQHFTRTQPVNTSAYTRV 60
+KN P R+ FDLS + FTAKVGELLP PGD +S +FTRT P+ ++A+TR+
Sbjct 13 LKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPLQSNAFTRL 72
Query 61 REYYDWFWCPLHLLWRNAPEVIAQIQQNVQH------ASSYDGSVLLGSNMPCVslsqls 114
RE +F+ P LW+ + + +N ASS G+ + + MPCV+ L
Sbjct 73 RENVQYFFVPYSALWKYFDSQVLNMTKNANGGDISRIASSLVGNQKVTTQMPCVNYKTLH 132
Query 115 kllsslkgkkNY-----------FGFDRSDLAYKILQYLRYGNV----------QTSSST 153
L + G R + K+LQ L YGN +
Sbjct 133 AYLLKFINRSTVGSDGSVGPEFNRGCYRHAESAKLLQLLGYGNFPEQFANFKVNNDKHNQ 192
Query 154 SGKNFGTSIPLSDRSYSQDYVFNHA--LSIFPLLGYKKFCQDYFRFTQWQDSAPYLWNID 211
SG+NF +D +N++ LSIF LL Y K C D++ + QWQ L N+D
Sbjct 193 SGQNF------------KDVTYNNSPYLSIFRLLAYHKICNDHYLYRQWQPYNASLCNVD 240
Query 212 YYDAKKST---------SILPDTFSTSYLTHNTLIDMEYCNWNKDMFFGVLPDAQYGDAS 262
Y S+ SI D+ L L+DM + N D F GVLP +Q+G S
Sbjct 241 YLTPNSSSLLSIDDALLSIPDDSIKAEKLN---LLDMRFSNLPLDYFTGVLPTSQFGSES 297
Query 263 VVDISFG-MSGQTVV-ASPSDISSRYTISNP--------SDSSTPNL------------- 299
VV+++ G SG V+ + S S R+ + + S+ NL
Sbjct 298 VVNLNLGNASGSAVLNGTTSKDSGRWRTTTGEWEMEQRVASSANGNLKLDNSNGTFISHD 357
Query 300 ---SGSPLV-------LDVLALRRGEALQRFREISLCTPANYRSQIKAHFGVDVGSELSG 349
SG+ + L ++ALR A Q+++EI L +++SQ++AHFG+ E +
Sbjct 358 HTFSGNVAINTSLSGNLSIIALRNALAAQKYKEIQLANDVDFQSQVEAHFGIKP-DEKNE 416
Query 350 MSTYIGGEASSLDISEVVNTNITESNEALIAGKGIGTGQFSDKFYAKDWGILMCIYHSVP 409
S +IGG +S ++I+E +N N++ N+A G G S KF AK +G+++ IY P
Sbjct 417 NSLFIGGSSSMININEQINQNLSGDNKATYGAAPQGNGSASIKFTAKTYGVVIGIYRCTP 476
Query 410 LLDYVLTSPDPQLFLSENTSFPVPELDAIGLESIPLSC-------YSNSSLEIPITNPNV 462
+LD+ D LF ++ + F +PE+D+IG++ C Y++ + + +
Sbjct 477 VLDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQT-FRCEVAAPAPYNDEFKAFRVGDGSS 535
Query 463 DAASLTMGYLPRYYAWKTSLDYVLGAFTTTEKEWVAPITASLWSKMLLPVTVDGSGINY- 521
S T GY PRY +KTS D GAF + K WV I + + V +GIN
Sbjct 536 PDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGIN---FDAIQNNVWNTWAGINAP 592
Query 522 NFFKVNPSILDPIFLVNADSTWDTDTFLVNAAFDIRVARNLDYDGMPY 569
N F P I+ +FLV++ + D D V RNL G+PY
Sbjct 593 NMFACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYATRNLSRYGLPY 640
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 187 bits (475), Expect = 1e-48, Method: Compositional matrix adjust.
Identities = 170/581 (29%), Positives = 255/581 (44%), Gaps = 71/581 (12%)
Query 3 NHPRRSGFDLSSKVAFTAKVGELLPVKWCLTMPGDKFSLSEQHFTRTQPVNTSAYTRVRE 62
N PR S FDLS K +TA G LLPV M D + Q F RT P+N++A+ +R
Sbjct 15 NRPR-SAFDLSQKHLYTAPAGALLPVLSVDLMFHDHIRIQAQDFMRTMPMNSAAFISMRG 73
Query 63 YYDWFWCPLHLLWRNAPEVIAQIQQ-NVQHASSYDGSVLLGSNMPCVslsqlskllsslk 121
Y++F+ P LW + I + SS G L S +P V +
Sbjct 74 VYEFFFVPYSQLWHPYDQFITSMNDYRSSVVSSAAGDKALDS-VPNV-KLADMYKFVRER 131
Query 122 gkkNYFGFDRSDLAYKILQYLRYGNVQTSSSTSGKNFGTSIPLSDRSYSQDYVFNHALSI 181
K+ FG+ S+ + +++ L YG TSS T +PL ++ +++
Sbjct 132 TDKDIFGYPHSNNSCRLMDLLGYGKPITSSK-------TPVPL---------LYTGNVNL 175
Query 182 FPLLGYKKFCQDYFRFTQWQDSAPYLWNIDYYDAKKSTSI-LPDTFSTSYLTHNTLIDME 240
F LL Y K DY+R T ++ Y +NID+ KK T + D F +++
Sbjct 176 FRLLAYNKIYSDYYRNTTYEGVDVYSFNIDH---KKGTFVPTADEFK-------KYLNLH 225
Query 241 YCNWNKDMFFGVLPDAQYGDASVVDISFGMSGQTVVASPSDISSRYTISNPSDSSTPNLS 300
Y N D + + P + + G + V SD + S +S+ N++
Sbjct 226 YRNAPLDFYTNLRPTPLF--------TIGSDSFSSVLQLSDPTGSAGFSADGNSAKLNMA 277
Query 301 GSPLVLDVLALRRGEALQRFREISLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASS 360
SP VL+V A+R AL + IS+ Y QI+AHFGV V G Y+GG S+
Sbjct 278 -SPDVLNVSAIRSAFALDKLLSISMRAGKTYAEQIEAHFGVTVSEGRDGQVYYLGGFDSN 336
Query 361 LDISEVVNT------NITESNEALIA-------GKGIGTGQFSDKFYAKDWGILMCIYHS 407
+ + +V T N++E A +A GKG G+G +F AK+ G+LMCIY
Sbjct 337 VQVGDVTQTSGTTNPNVSEVGNAKLAGYLGKITGKGTGSGYGEIQFDAKEPGVLMCIYSV 396
Query 408 VPLLDYVLTSPDPQLFLSENTSFPVPELDAIGLESIPLSCYSNSSLEIPITNPNVDAASL 467
VP + Y DP + + +PE + +G++ I +P A
Sbjct 397 VPAMQYDCMRLDPFVAKQTRGDYFIPEFENLGMQPI-----------VPAFVSLNRAKDN 445
Query 468 TMGYLPRYYAWKTSLDYVLGAFTTTEKEWVAPITASLWSKMLLPVTVDGSGINYNFFKVN 527
+ G+ PRY +KT+ D G F E P+ S WS + + N K+N
Sbjct 446 SYGWQPRYSEYKTAFDINHGQFANGE-----PL--SYWSIARARGSDTLNTFNVAALKIN 498
Query 528 PSILDPIFLVNADSTWDTDTFLVNAAFDIRVARNLDYDGMP 568
P LD +F VN + T TD A F+I ++ DGMP
Sbjct 499 PHWLDSVFAVNYNGTEVTDCMFGYAHFNIEKVSDMTEDGMP 539
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 173 bits (439), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 163/583 (28%), Positives = 259/583 (44%), Gaps = 69/583 (12%)
Query 7 RSGFDLSSKVAFTAKVGELLPVKWCLTMPGDKFSLSEQHFTRTQPVNTSAYTRVREYYDW 66
R+ FDLS + FTA G LLPV +P D ++ Q F RT P+NT+A+ +R Y++
Sbjct 17 RNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAFASMRGVYEF 76
Query 67 FWCPLHLLWRNAPEVIAQIQQNVQHASSYDGSVLLGSNMPCVslsqlskllsslkgkkNY 126
F+ P H LW + I + N H+S+ + S+ G++ V + + +SL K
Sbjct 77 FFVPYHQLWAQFDQFITGM--NDFHSSA-NKSIQGGTSPLQVPYFNVDSVFNSLNTGKES 133
Query 127 FGFDRSDLAYK-------ILQYLRYGNVQTSSSTSGKNFGTSIPLSDRSYSQDYVFNHAL 179
DL YK +L L YG S FGT+ P + + +N
Sbjct 134 GSGSTDDLQYKFKYGAFRLLDLLGYGRKFDS-------FGTAYPDNVSGLKNNLDYN--C 184
Query 180 SIFPLLGYKKFCQDYFRFTQWQDSAPYLWNIDYY-----DAKKSTSILPDTFSTSYLTHN 234
S+F +L Y K QDY+R + +++ +N D + DAK ++ D F Y N
Sbjct 185 SVFRILAYNKIYQDYYRNSNYENFDTDSFNFDKFKGGLVDAK----VVADLFKLRY--RN 238
Query 235 TLIDMEYCNWNKDMFFGVLPDAQYGDASVVDISFGMSGQTVVASPSDISSRYTISNPSDS 294
D + N + F + D ++I + + V S +R +DS
Sbjct 239 AQTDY-FTNLRQSQLFSFT--TAFEDVDNINI----APRDYVKSDGSNFTRVNFGVDTDS 291
Query 295 STPNLSGSPLVLDVLALRRGEALQRFREISLCTPANYRSQIKAHFGVDVGSELSGMSTYI 354
S + S V +LR A+ + +++ ++ Q++AH+GV++ G Y+
Sbjct 292 SEGDFS-------VSSLRAAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYL 344
Query 355 GGEASSLDISEVVNTNITESNE--------ALIAGKGIGTGQFSDKFYAKDWGILMCIYH 406
GG S + +S+V T+ T + E +AGKG G+G+ F AK+ G+LMCIY
Sbjct 345 GGFDSDMQVSDVTQTSGTTATEYKPEAGYLGRVAGKGTGSGRGRIVFDAKEHGVLMCIYS 404
Query 407 SVPLLDYVLTSPDPQLFLSENTSFPVPELDAIGLESIPL-SCYSNSSLEIPITNPNVDAA 465
VP + Y T DP + + + PE + +G++ PL S Y +S NP
Sbjct 405 LVPQIQYDCTRLDPMVDKLDRFDYFTPEFENLGMQ--PLNSSYISSFCTTDPKNP----- 457
Query 466 SLTMGYLPRYYAWKTSLDYVLGAFTTTEKEWVAPITASLWSKMLLPVTVDGSGINYNFFK 525
+GY PRY +KT+LD G F ++ S WS + FK
Sbjct 458 --VLGYQPRYSEYKTALDVNHGQFAQSD-------ALSSWSVSRFRRWTTFPQLEIADFK 508
Query 526 VNPSILDPIFLVNADSTWDTDTFLVNAAFDIRVARNLDYDGMP 568
++P L+ IF V+ + T D F+I ++ DGMP
Sbjct 509 IDPGCLNSIFPVDYNGTEANDCVYGGCNFNIVKVSDMSVDGMP 551
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 162 bits (410), Expect = 9e-40, Method: Compositional matrix adjust.
Identities = 164/600 (27%), Positives = 266/600 (44%), Gaps = 61/600 (10%)
Query 5 PR--RSGFDLSSKVAFTAKVGELLPVKWCLTMPGDKFSLSEQHFTRTQPVNTSAYTRVRE 62
PR R+GFDLSS+ F+AK G+LLP+ P + F S Q RT +NT++Y R++E
Sbjct 6 PRLARNGFDLSSRRIFSAKAGQLLPIGCWEVNPSEHFKFSVQDLVRTTTLNTASYARMKE 65
Query 63 YYDWFWCPLHLLWRNAPEVIAQIQQNVQHASSYDGSVLLGS---NMPCVslsq---lskl 116
YY +F+ LW+ + I + N H S+ +G G+ N C S+ +
Sbjct 66 YYHFFFVSYRSLWQWFDQFI--VGTNNPH-SALNGVKKNGTTNYNQICSSVPTFDLGKLI 122
Query 117 lsslkgkkNYFGFDRSDLAYKILQYLRYGNVQTSSSTSGKNFGTS---IPLSDRSYSQDY 173
+ GF+ S+ A K+L L YG + +N TS +P D
Sbjct 123 TRLKTSDMDSQGFNYSEGAAKLLNMLNYGVTNKGKFMNLENLITSTSYLPSKDDKEPSS- 181
Query 174 VFNHALSIFPLLGYKKFCQDYFRFTQWQDSAPYLWNIDYYDAKKSTSILPDTFSTSYLTH 233
++ +S F LL Y+K D++R W S +N+D Y + +I PD
Sbjct 182 IYACKVSPFRLLAYQKIFNDFYRNQDWTPSDVRSFNVDDYADDSNLTIEPDVALK----- 236
Query 234 NTLIDMEYCNWNKDMFFGVLPDAQYGDASVVDISFGMSGQTVVASPSDISSRYTISNPSD 293
M Y + KD + P Y D + ++ + G V ++ S ++ D
Sbjct 237 --FCQMRYRPYAKDWLTSMKPTPNYSDG-IFNLPEYVRGNGNVILTNNKSGSVSL----D 289
Query 294 SSTPNLSGSPLVLDVLALRRGEALQRFREISL-CTPANYRSQIKAHFGVDVGSELSGMST 352
S T SP V LR AL + E + +Y SQI+AHFG V + +
Sbjct 290 SGTV----SPSSFSVNDLRAAFALDKMLEATRRANGLDYASQIEAHFGFKVPESRANDAR 345
Query 353 YIGGEASSLDISEVVNTNITESNEAL------IAGKGIGT-GQFSDKFYAKDWGILMCIY 405
++GG +S+ +SEVV+TN +++ + GKGIG+ + +F + + GI+MCIY
Sbjct 346 FLGGFDNSIVVSEVVSTNGNAASDGSHASIGDLGGKGIGSMSSGTIEFDSTEHGIIMCIY 405
Query 406 HSVPLLDYVLTSPDPQLFLSENTSFPVPELDAIGLESI---PLSCYSNSSLEIPITNPNV 462
P +Y + DP F PE +G +++ L C + E ++
Sbjct 406 SVAPQSEYNASYLDPFNRKLTREQFYQPEFADLGYQALIGSDLICSTLGMNEKQAGFSDI 465
Query 463 DAASLTMGYLPRYYAWKTSLDYVLGAFTTTE--KEWVAP---ITASLWSKMLLPVTVDGS 517
+ + +GY RY +KT+ D V G F + + W P K + P G+
Sbjct 466 ELNNNLLGYQVRYNEYKTARDLVFGDFESGKSLSYWCTPRFDFGYGDTEKKIAPENKGGA 525
Query 518 GI----------NYNFFKVNPSILDPIFLVNADSTWDTDTFLVNAAFDIRVARNLDYDGM 567
+ NF+ +NP++++PIFL +A D F+VN+ D++ R + G+
Sbjct 526 DYRKKGNRSHWSSRNFY-INPNLVNPIFLTSA---VQADHFIVNSFLDVKAVRPMSVTGL 581
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 159 bits (402), Expect = 5e-39, Method: Compositional matrix adjust.
Identities = 152/565 (27%), Positives = 248/565 (44%), Gaps = 70/565 (12%)
Query 25 LLPVKWCLTMPGDKFSLSEQHFTRTQPVNTSAYTRVREYYDWFWCPLHLLWRNAPEVIAQ 84
LLPV +P D ++ Q F RT P+NT+A+ +R Y++F+ P H LW + I
Sbjct 2 LLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAFASMRGVYEFFFVPYHQLWAQFDQFITG 61
Query 85 IQQNVQHASSYDGSVLLGS--------NMPCVslsqlskllsslkgkkNYFGFDRSDLAY 136
+ N H+S+ + S+ G+ N+ V + + + + + F A+
Sbjct 62 M--NDFHSSA-NKSIQGGTSPLQVPYFNLESVFKNIIERDSTPSFQDDLQYRFKYG--AF 116
Query 137 KILQYLRYGNVQTSSSTSGKNFGTSIPLSDRSYSQDYVFNHALSIFPLLGYKKFCQDYFR 196
++L L YG S FGT+ P + + +N S+F +L Y K QDY+R
Sbjct 117 RLLDLLGYGRKFDS-------FGTAYPDNVSGLKNNLDYN--CSVFRVLAYNKIYQDYYR 167
Query 197 FTQWQDSAPYLWNIDYY-----DAKKSTSILPDTFSTSYLTHNTLIDMEYCNWNKDMFFG 251
+ +++ +N D + DAK ++ D F Y N D + N + F
Sbjct 168 NSNYENFDTDSFNFDKFKGGLVDAK----VVADLFKLRY--RNAQTDY-FTNLRQSQLFT 220
Query 252 VLPDAQYGDASVVDISFGMSGQTVVASPSDISSRYTISNPSDSSTPNLSGSPLVLDVLAL 311
+P ++ D ++ Q S S+ + ++ P D NL V +L
Sbjct 221 FIP--EFSDDEHLNFD---RDQYADQSKSNFTQ---LNFPVDVDN-NLG----YFSVSSL 267
Query 312 RRGEALQRFREISLCTPANYRSQIKAHFGVDVGSELSGMSTYIGGEASSLDISEVVNTNI 371
R A+ + +++ ++ Q++AH+GV++ G Y+GG S L +S+V T+
Sbjct 268 RSAFAVDKLLSVTMRAGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDLQVSDVTQTSG 327
Query 372 TESNE--------ALIAGKGIGTGQFSDKFYAKDWGILMCIYHSVPLLDYVLTSPDPQLF 423
T + E IAGKG G+G+ F AK+ G+LMCIY VP + Y T DP +
Sbjct 328 TTATEYKPEAGYLGRIAGKGTGSGRGRIVFDAKEHGVLMCIYSLVPQIQYDCTRLDPMVD 387
Query 424 LSENTSFPVPELDAIGLESIPLSCYSNSSLEIPITNPNVDAASLTMGYLPRYYAWKTSLD 483
+ F PE + +G++ PL+ SS P D + +GY PRY +KT+LD
Sbjct 388 KLDRFDFFTPEFENLGMQ--PLNSSYISSFCTP------DPKNPVLGYQPRYSEYKTALD 439
Query 484 YVLGAFTTTEKEWVAPITASLWSKMLLPVTVDGSGINYNFFKVNPSILDPIFLVNADSTW 543
G F + S WS + FK++P L+ +F V + T
Sbjct 440 INHGQFAQND-------ALSSWSVSRFRRWTTFPQLEIADFKIDPGCLNSVFPVEFNGTE 492
Query 544 DTDTFLVNAAFDIRVARNLDYDGMP 568
TD F+I ++ DGMP
Sbjct 493 STDCVFGGCNFNIVKVSDMSVDGMP 517
Lambda K H a alpha
0.319 0.134 0.414 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4156463374755