bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_6
Length=369
Score E
Sequences producing significant alignments: (Bits) Value
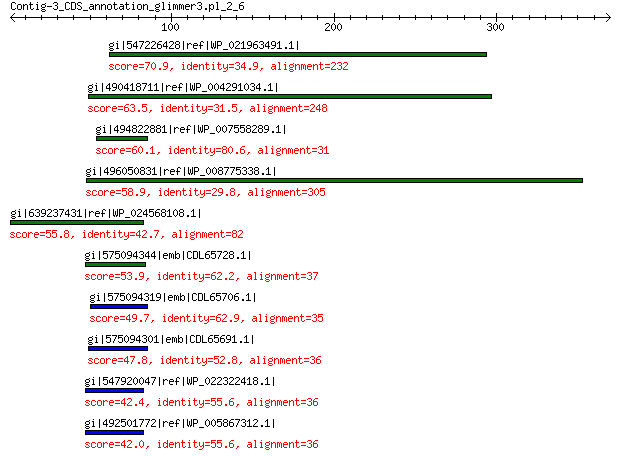
gi|547226428|ref|WP_021963491.1| putative uncharacterized protein 70.9 3e-10
gi|490418711|ref|WP_004291034.1| hypothetical protein 63.5 7e-08
gi|494822881|ref|WP_007558289.1| hypothetical protein 60.1 9e-07
gi|496050831|ref|WP_008775338.1| predicted protein 58.9 2e-06
gi|639237431|ref|WP_024568108.1| hypothetical protein 55.8 2e-05
gi|575094344|emb|CDL65728.1| unnamed protein product 53.9 9e-05
gi|575094319|emb|CDL65706.1| unnamed protein product 49.7 0.002
gi|575094301|emb|CDL65691.1| unnamed protein product 47.8 0.009
gi|547920047|ref|WP_022322418.1| putative uncharacterized protein 42.4 0.29
gi|492501772|ref|WP_005867312.1| hypothetical protein 42.0 0.43
>gi|547226428|ref|WP_021963491.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103380|emb|CCY83991.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=416
Score = 70.9 bits (172), Expect = 3e-10, Method: Compositional matrix adjust.
Identities = 81/274 (30%), Positives = 126/274 (46%), Gaps = 60/274 (22%)
Query 62 YNSASAQRSRLEEAGLNPYLMMNggsagtaqssgsstpas-----------assplsMQ- 109
YNSA AQR+RLE AGLNPYLMM+GG+AG + S+ + +S MQ
Sbjct 91 YNSAKAQRARLEAAGLNPYLMMSGGNAGAVSAVSGSSGSGGSPSPMGVNPPTASSAVMQA 150
Query 110 -RQDFSGLSNTLASALQISNQTKETNANVQTLQSQKSLYDAQANSILSNVDWWKLGPEYK 168
R DFSG++ + + L I Q+QK + DAQA S+ +K+ +YK
Sbjct 151 FRPDFSGVTGIIQTLLDI--------------QAQKGVRDAQAFSLGEQASGFKIENKYK 196
Query 169 -----------------KWSQMT----GLARAGLQFQTD----KQNLRNMTWSGNLIQAQ 203
K SQ + AR F +D ++ N ++G LI+AQ
Sbjct 197 AEKLLWDIYNSKADYNLKNSQESLNNMSFARLQAMFSSDVSKAQREAENAQFTGELIRAQ 256
Query 204 ----HIGALLDNKSKRIINNYLDEGQRLQLDLMAAQYYDAMASGHLKYQQAKSEITKRIL 259
+ LL K + Y D+ +L +M+AQ Y +A+G QA+ I +
Sbjct 257 TACQQLQGLLGAKELK----YYDQKVLQELAIMSAQQYSLVAAGKASEAQARQAIENALN 312
Query 260 MMAEAKGLQINNKAAEDTADGYIKALNAEYSASY 293
++ + +G++++N + TA+ IK + SY
Sbjct 313 LVEQREGIKVDNYVKQKTANALIKTARNNCNTSY 346
>gi|490418711|ref|WP_004291034.1| hypothetical protein [Bacteroides eggerthii]
gi|217986638|gb|EEC52972.1| hypothetical protein BACEGG_02723 [Bacteroides eggerthii DSM
20697]
Length=368
Score = 63.5 bits (153), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 78/262 (30%), Positives = 120/262 (46%), Gaps = 16/262 (6%)
Query 49 RQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMMNggsagtaqssgsstpasassplsM 108
+ +Q +MWNK NEYN SAQR+RLE AGLNPY+MMNGGSAG A S + ++ S+
Sbjct 86 KAYQTEMWNKQNEYNDPSAQRARLEAAGLNPYMMMNGGSAGVAGSVSGTQGSAPSAGSPS 145
Query 109 QR-----------QDFSGLSNTLASALQISNQTKETNANVQTLQSQKSLYDAQANSILSN 157
+ D+SG+ L A I + N+Q Q+ + + + +
Sbjct 146 AQGVQPPTATPYSADYSGVMQGLGHA--IDTIMTGSQRNIQNAQADNLRIEGKYIASKAI 203
Query 158 VDWWKLGPEYKKWSQMTGLARAGLQFQTDKQNLRNMTWSGNL--IQAQHIGALLDNKSKR 215
+ +K E K + + R Q D + + N+ IQAQ A+ +N +
Sbjct 204 AELYKTYNEAKNDDERVAIQRVLSSIQKDLSASQVAVNNENVRQIQAQTKIAVTENLLRE 263
Query 216 IINNYLDEGQRLQLDLMAAQYYDAMASGHLKYQQAKSEITKRILMMAEAKGLQINNKAAE 275
+L QR QL L AA A +L +QA+ EI K + A G + N+
Sbjct 264 QQLKFLPYEQRTQLALGAADIALKYAQKNLTEKQARHEIEKLAETIVRANGQAMQNQYDA 323
Query 276 DTADGYIKALNAE-YSASYDIN 296
+T +K + ++A YD +
Sbjct 324 ETYRDRVKLVKESLFNAIYDTD 345
>gi|494822881|ref|WP_007558289.1| hypothetical protein [Bacteroides plebeius]
gi|198272097|gb|EDY96366.1| hypothetical protein BACPLE_00802 [Bacteroides plebeius DSM 17135]
Length=344
Score = 60.1 bits (144), Expect = 9e-07, Method: Compositional matrix adjust.
Identities = 25/31 (81%), Positives = 28/31 (90%), Gaps = 0/31 (0%)
Query 54 DMWNKTNEYNSASAQRSRLEEAGLNPYLMMN 84
DMWN NEYNSAS+QR RLEEAGLNPY+MM+
Sbjct 71 DMWNAENEYNSASSQRKRLEEAGLNPYMMMD 101
>gi|496050831|ref|WP_008775338.1| predicted protein [Bacteroides sp. 2_2_4]
gi|229448895|gb|EEO54686.1| hypothetical protein BSCG_01611 [Bacteroides sp. 2_2_4]
Length=381
Score = 58.9 bits (141), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 91/324 (28%), Positives = 150/324 (46%), Gaps = 46/324 (14%)
Query 48 ARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMMNggsagtaqssgsstpasasspls 107
A+Q DM+N TNEYNSASAQR R E AGLNPY+MMN GSAGTA ++ +++ + +
Sbjct 78 AKQNAWDMFNATNEYNSASAQRERYEAAGLNPYVMMNTGSAGTAAATSATSATAPTKQGI 137
Query 108 MQ------RQDFSGLSNTLASAL-QISN--QTKETNANVQTLQSQKSLYDAQANSILSNV 158
D+SG+ L A+ Q+S+ +T A L+ + A+A + ++N+
Sbjct 138 TPPTASPYSADYSGIMQGLGQAIDQLSSIPDKAKTIAETGNLKIEGKYKAAEAIARIANI 197
Query 159 DWWKLGPEYKK---------WSQMTGLARAGLQFQTDKQNLRNMTWSGNLIQAQHIGALL 209
K KK +S LA + + + QN+ NM ++I L+
Sbjct 198 ---KADTHSKKEQVALNKLMYSIQKDLASSTM--AVNSQNIANMRAEEKF---KNIQTLI 249
Query 210 DNKSKRIINNYLDEGQRLQLDLMAAQYYDAMASGHLKYQQAKSEITKRILMMAEAKGLQI 269
+K +++D Q+++L AA +A G L QA EI K + EA+ I
Sbjct 250 ADKQL----SFMDATQKMELAEKAANIQLKLAQGALTRNQAAHEIKK--ISETEARTTLI 303
Query 270 NNKAAEDTADGYIKALNAEYSASYDINSPFKYGEDSY-VPASVLKSRMDALNSKWQFDKR 328
N + + I+ + + ++ D+Y V L+ + F+
Sbjct 304 NEQTSLT-----IEQNTGQQLQNQAQRQQNRFDADTYNVRVKTLEESL--------FNIV 350
Query 329 YWTEGLNALGTVGNAVGSVGNLRK 352
+ T+ L A+ TVG + +VG++ K
Sbjct 351 FETDKLGAVKTVGKGIRAVGSVAK 374
>gi|639237431|ref|WP_024568108.1| hypothetical protein [Elizabethkingia anophelis]
Length=287
Score = 55.8 bits (133), Expect = 2e-05, Method: Compositional matrix adjust.
Identities = 35/82 (43%), Positives = 39/82 (48%), Gaps = 18/82 (22%)
Query 1 MGLLDFIPVVGDIASSIGNVVSTnkannanmavnrmnnefnaaeaeKARQFQLDMWNKTN 60
M LL IPVVG I N S K + R F LDMWN+ N
Sbjct 1 MSLLSAIPVVGGIIEGAMNNSSNKKIAR------------------ENRAFALDMWNRNN 42
Query 61 EYNSASAQRSRLEEAGLNPYLM 82
EYN+ AQ RL+EAGLNP LM
Sbjct 43 EYNTPLAQMQRLKEAGLNPNLM 64
>gi|575094344|emb|CDL65728.1| unnamed protein product [uncultured bacterium]
Length=368
Score = 53.9 bits (128), Expect = 9e-05, Method: Compositional matrix adjust.
Identities = 23/37 (62%), Positives = 26/37 (70%), Gaps = 0/37 (0%)
Query 47 KARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMM 83
K R FQLDMWN+ NEYN Q RLEEAG+NP+ M
Sbjct 59 KQRDFQLDMWNRNNEYNKPDEQMKRLEEAGINPWQSM 95
>gi|575094319|emb|CDL65706.1| unnamed protein product [uncultured bacterium]
Length=396
Score = 49.7 bits (117), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 22/35 (63%), Positives = 25/35 (71%), Gaps = 0/35 (0%)
Query 50 QFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMMN 84
+F MWN NEYN QR+RLE AGLNPYLMM+
Sbjct 72 KFNERMWNLQNEYNRPDMQRARLEAAGLNPYLMMD 106
>gi|575094301|emb|CDL65691.1| unnamed protein product [uncultured bacterium]
Length=437
Score = 47.8 bits (112), Expect = 0.009, Method: Compositional matrix adjust.
Identities = 19/36 (53%), Positives = 26/36 (72%), Gaps = 0/36 (0%)
Query 49 RQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLMMN 84
R F MW TN+YN+ AQ+ RLE+AG+NPY+ M+
Sbjct 63 RDFARQMWKDTNDYNTPIAQKQRLEQAGMNPYVNMD 98
>gi|547920047|ref|WP_022322418.1| putative uncharacterized protein [Parabacteroides merdae CAG:48]
gi|524592959|emb|CDD13571.1| putative uncharacterized protein [Parabacteroides merdae CAG:48]
Length=259
Score = 42.4 bits (98), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 20/36 (56%), Positives = 26/36 (72%), Gaps = 0/36 (0%)
Query 47 KARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLM 82
KA ++MWN N+YNS +AQ SRL +AGLNP L+
Sbjct 17 KAYARSVEMWNMQNQYNSPTAQMSRLRQAGLNPNLV 52
>gi|492501772|ref|WP_005867312.1| hypothetical protein [Parabacteroides distasonis]
gi|409230405|gb|EKN23269.1| hypothetical protein HMPREF1059_03254 [Parabacteroides distasonis
CL09T03C24]
Length=288
Score = 42.0 bits (97), Expect = 0.43, Method: Compositional matrix adjust.
Identities = 20/36 (56%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 47 KARQFQLDMWNKTNEYNSASAQRSRLEEAGLNPYLM 82
KA Q L+MWN NEYNS + Q +R+ AGLNP L+
Sbjct 52 KAYQRSLNMWNLQNEYNSPTQQMARIRAAGLNPNLV 87
Lambda K H a alpha
0.313 0.127 0.365 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2257338701640