bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-3_CDS_annotation_glimmer3.pl_2_1
Length=233
Score E
Sequences producing significant alignments: (Bits) Value
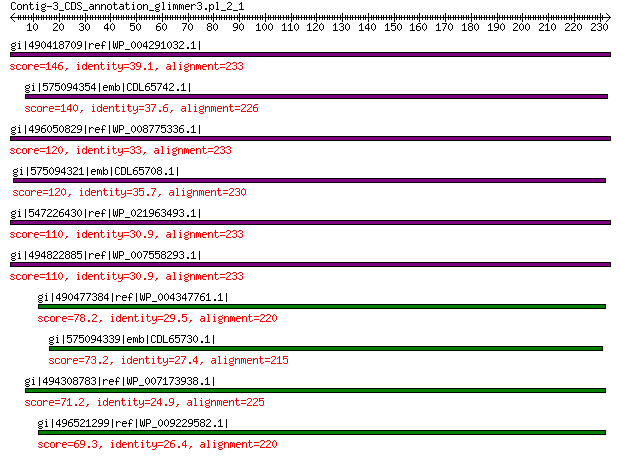
gi|490418709|ref|WP_004291032.1| hypothetical protein 146 5e-37
gi|575094354|emb|CDL65742.1| unnamed protein product 140 5e-35
gi|496050829|ref|WP_008775336.1| hypothetical protein 120 8e-28
gi|575094321|emb|CDL65708.1| unnamed protein product 120 2e-27
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 110 3e-24
gi|494822885|ref|WP_007558293.1| hypothetical protein 110 3e-24
gi|490477384|ref|WP_004347761.1| capsid protein 78.2 3e-13
gi|575094339|emb|CDL65730.1| unnamed protein product 73.2 1e-11
gi|494308783|ref|WP_007173938.1| hypothetical protein 71.2 4e-11
gi|496521299|ref|WP_009229582.1| capsid protein 69.3 2e-10
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 146 bits (368), Expect = 5e-37, Method: Compositional matrix adjust.
Identities = 91/238 (38%), Positives = 126/238 (53%), Gaps = 35/238 (15%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
MA+ LK ++N+P ++G D+S K FTAK GELLPV +P T++INL+ FTRT+P
Sbjct 1 MANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAP-IQSKDLLTNLTVKGDLPYCS 119
V T+A+ RIREY+DFF VP DL+W ++ + QM + S D N + G++PY +
Sbjct 61 VNTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDNPQHAVSIDPTRNFVLSGEMPYMT 120
Query 120 LSDLSFALKFASGNPASLGNKVSVSPGFG----NIFGYNRGDVNHKLLSMLDYGNVVDKS 175
+ AS N +S + N FGYNR + KLL L YGN
Sbjct 121 SEAI-----------ASYINALSTASALADYKSNYFGYNRSKSSVKLLEYLGYGNY---- 165
Query 176 ANWIGTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEKC 233
+++ + WN A NL N+F L YQKIY DF+R SQWE+
Sbjct 166 ESFLTDD----WNTAPLMA-----------NLNHNIFGLLAYQKIYSDFYRDSQWERV 208
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 140 bits (353), Expect = 5e-35, Method: Compositional matrix adjust.
Identities = 85/227 (37%), Positives = 122/227 (54%), Gaps = 30/227 (13%)
Query 7 LKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAY 66
+ +++NRP ++G D+S K FTAK GELLPV T +P ++ INL FTRT+P+ TSA+
Sbjct 3 MADIKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTSAF 62
Query 67 TRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLL-TNLTVKGDLPYCSLSDLSF 125
R+REY+DF+ VP + +W FDS + QM S L N + G +PY + ++
Sbjct 63 ARMREYYDFYFVPFEQMWNKFDSCITQMNANVQHASGPTLDDNTPLSGRMPYFTSEQIAD 122
Query 126 ALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANR 185
L + A+ K N FG+NR + KLL L YG+ N + N
Sbjct 123 YLN----DQATAARK--------NPFGFNRSTLTCKLLQYLGYGDY-----NSFDSETNT 165
Query 186 WWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEK 232
W +A +Y NL ++ F L YQKIY DF+R++QWEK
Sbjct 166 W------SAKPLLY------NLELSPFPLLAYQKIYSDFYRYTQWEK 200
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 120 bits (301), Expect = 8e-28, Method: Compositional matrix adjust.
Identities = 77/235 (33%), Positives = 122/235 (52%), Gaps = 25/235 (11%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
MA+ LK L+N+ ++G D+S+K FTAK GELLPV +P + I+L+ FTRT+P
Sbjct 1 MANIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEVLPGDKWSIDLKSFTRTQP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLL--TNLTVKGDLPYC 118
+ T+A+ R+REY+DF+ VP +L+W ++ + QM + P + + N + G +P
Sbjct 61 LNTAAFARMREYYDFYFVPYNLLWNKANTVLTQMYDN-PQHATSYIPSANQALAGVMPNV 119
Query 119 SLSDLSFALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANW 178
+ ++ L + + + + N FGY+R KLL L YGN +
Sbjct 120 TCKGIADYLNLVAPDVTTTNSYEK------NYFGYSRSLGTAKLLEYLGYGNFYTYA--- 170
Query 179 IGTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEKC 233
T+ N W + ++ NL +N++ + YQKIY D R SQWEK
Sbjct 171 --TSKNNTWTKSPLSS-----------NLQLNIYGVLAYQKIYADHIRDSQWEKV 212
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 120 bits (300), Expect = 2e-27, Method: Compositional matrix adjust.
Identities = 82/241 (34%), Positives = 128/241 (53%), Gaps = 26/241 (11%)
Query 2 AHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPV 61
++ GL L+N+P ++ D+S +N FTAKVGELLP + P + +++ YFTRT P+
Sbjct 5 SNIMGLHGLKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTRTAPL 64
Query 62 QTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQM------GEKAPIQSKDLLTNLTVKGDL 115
Q++A+TR+RE +F VP +WK FDS V+ M G+ + I S L+ N V +
Sbjct 65 QSNAFTRLRENVQYFFVPYSALWKYFDSQVLNMTKNANGGDISRIASS-LVGNQKVTTQM 123
Query 116 PYCSLSDL-SFALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNH----KLLSMLDYGN 170
P + L ++ LKF N +++G+ SV P F NRG H KLL +L YGN
Sbjct 124 PCVNYKTLHAYLLKFI--NRSTVGSDGSVGPEF------NRGCYRHAESAKLLQLLGYGN 175
Query 171 VVDKSANWIGTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQW 230
++ AN+ + N + + YN + +++F L Y KI D + + QW
Sbjct 176 FPEQFANF------KVNNDKHNQSGQNFKDVTYNNSPYLSIFRLLAYHKICNDHYLYRQW 229
Query 231 E 231
+
Sbjct 230 Q 230
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 110 bits (275), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 72/237 (30%), Positives = 121/237 (51%), Gaps = 37/237 (16%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
M+ L L+N ++G D+S KN FTAKVGELLP+ P + I + FTRT+P
Sbjct 1 MSSVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQP 60
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSL 120
V ++AY+R+REY+DF+ VP L+W + M + P + DL++++ + P+ +
Sbjct 61 VNSAAYSRLREYYDFYFVPYRLLWNMAPTFFTNMPD--PHHAADLVSSVNLSQRHPWFTF 118
Query 121 SDLSFALKFASGNPASLGNKVSVSPGF----GNIFGYNRGDVNHKLLSMLDYGNVVDKSA 176
D+ LGN S+S + N FG++R +++ KLL+ L+YG D +
Sbjct 119 FDIM----------EYLGNLNSLSGAYEKYQKNFFGFSRVELSVKLLNYLNYGFGKDYES 168
Query 177 NWIGTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEKC 233
+ ++++ ++ ++ F L YQKI +D+FR QW+
Sbjct 169 VKVPSDSD---------------------DIVLSPFPLLAYQKICEDYFRDDQWQSA 204
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 110 bits (275), Expect = 3e-24, Method: Compositional matrix adjust.
Identities = 72/234 (31%), Positives = 112/234 (48%), Gaps = 27/234 (12%)
Query 1 MAHFTGLKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRP 60
MA+ +K ++N+P ++G+D++ K FTAK G L+PVW +P ++ F RT+P
Sbjct 8 MANIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQP 67
Query 61 VQTSAYTRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLT-NLTVKGDLPYCS 119
+ T+A+ R+R YFDF+ VP +W F +A+ QM S +L N+ + +LPY +
Sbjct 68 LNTAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLADNVPLSDELPYFT 127
Query 120 LSDLSFALKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWI 179
++ + SL + N FGY R + +L L YG+
Sbjct 128 AEQVADYI-------VSLADS-------KNQFGYYRAWLVCIILEYLGYGDFYPYIVEAA 173
Query 180 GTNANRWWNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWEKC 233
G W R + NL + F L YQKIY DF R++QWE+
Sbjct 174 GGEGATWATRPMLN------------NLKFSPFPLFAYQKIYADFNRYTQWERS 215
>gi|490477384|ref|WP_004347761.1| capsid protein [Prevotella buccalis]
gi|281300712|gb|EFA93043.1| putative capsid protein (F protein) [Prevotella buccalis ATCC
35310]
Length=552
Score = 78.2 bits (191), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 65/220 (30%), Positives = 100/220 (45%), Gaps = 25/220 (11%)
Query 12 NRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAYTRIRE 71
NRP ++ D+S K+ FTA G LLPV T IP+ I F R P+ ++A+ +R
Sbjct 14 NRP-RNAFDLSQKHLFTAHAGMLLPVMTLDLIPHDHVSIQATDFMRCLPMNSAAFMSMRS 72
Query 72 YFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSLSDLSFALKFAS 131
++FF VP +W FD + M + + DL + K L S F +
Sbjct 73 VYEFFFVPYSQLWHPFDQFITGMNDYRSVLQSDLYKS---KSPLVIPSFKRKELYELFNA 129
Query 132 GNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANRWWNRRV 191
P N+ S P +IFG+ +LL +L YG V NA+
Sbjct 130 --PGGFLNQQSNQP---DIFGFKSRFNFLRLLDLLGYGVYV---------NAD------- 168
Query 192 ATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWE 231
++ +S+ + +++F LA YQKIY DF+R + +E
Sbjct 169 GSSRIDAFSKLLDDTEKLSIFRLAAYQKIYSDFYRNTTYE 208
>gi|575094339|emb|CDL65730.1| unnamed protein product [uncultured bacterium]
Length=588
Score = 73.2 bits (178), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 59/217 (27%), Positives = 108/217 (50%), Gaps = 36/217 (17%)
Query 16 KSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAYTRIREYFDF 75
K+G D+S ++ FT+ VG+LLPV+ D+ P RI+ FTRT+P++++A R+ E+ ++
Sbjct 16 KNGFDMSQRHPFTSSVGQLLPVFYDYLNPGDKIRISANLFTRTQPMKSTAMARLTEHIEY 75
Query 76 FAVPCDLIWKSFDSAVIQMGEKAPIQSKDLL--TNLTVKGDLPYCSLSDLSFALKFASGN 133
F VP + ++ F S + + S L+ NLT +P+ +S AL+ A +
Sbjct 76 FFVPFEQMFSLFGSVFYGIDD---YNSSSLVKHNNLT----MPFFKSDAVSAALEAAYTS 128
Query 134 PASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANRWWNRRVAT 193
+S N+ ++P ++ G R +L ML YG+++ + N + +A+
Sbjct 129 FSSSINRKVLTP---DMMGQPRVYGILRLSEMLGYGSLLLSNDNNLLPHAD--------- 176
Query 194 ADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQW 230
+++F YQKI+ DF+R +
Sbjct 177 ---------------MSVFLFTAYQKIFNDFYRLDDY 198
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 71.2 bits (173), Expect = 4e-11, Method: Compositional matrix adjust.
Identities = 56/225 (25%), Positives = 97/225 (43%), Gaps = 26/225 (12%)
Query 7 LKELQNRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAY 66
+K + +++ D+S ++ FTA G LLPV IP+ IN + F RT P+ T+A+
Sbjct 8 IKATRPNRNRNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAF 67
Query 67 TRIREYFDFFAVPCDLIWKSFDSAVIQMGEKAPIQSKDLLTNLTVKGDLPYCSLSDLSFA 126
+R ++FF VP +W FD + M + +K + T +PY ++ + +
Sbjct 68 ASMRGVYEFFFVPYHQLWAQFDQFITGMNDFHSSANKSIQGG-TSPLQVPYFNVDSVFNS 126
Query 127 LKFASGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANRW 186
L + + + + +G +LL +L YG D N
Sbjct 127 LNTGKESGSGSTDDLQYKFKYGAF----------RLLDLLGYGRKFDSFGTAYPDN---- 172
Query 187 WNRRVATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWE 231
V K N++ ++F + Y KIYQD++R S +E
Sbjct 173 -----------VSGLKNNLDYNCSVFRILAYNKIYQDYYRNSNYE 206
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 69.3 bits (168), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 58/221 (26%), Positives = 98/221 (44%), Gaps = 41/221 (19%)
Query 12 NRPHKSGHDISAKNCFTAKVGELLPVWTDFAIPNCTYRINLEYFTRTRPVQTSAYTRIRE 71
NRP +S D+S K+ +TA G LLPV + + + RI + F RT P+ ++A+ +R
Sbjct 15 NRP-RSAFDLSQKHLYTAPAGALLPVLSVDLMFHDHIRIQAQDFMRTMPMNSAAFISMRG 73
Query 72 YFDFFAVPCDLIWKSFDSAVIQMGE-KAPIQSKDLLTNLTVKGDLPYCSLSDLSFALKFA 130
++FF VP +W +D + M + ++ + S + GD S+ ++ A +
Sbjct 74 VYEFFFVPYSQLWHPYDQFITSMNDYRSSVVS-------SAAGDKALDSVPNVKLADMY- 125
Query 131 SGNPASLGNKVSVSPGFGNIFGYNRGDVNHKLLSMLDYGNVVDKSANWIGTNANRWWNRR 190
K +IFGY + + +L+ +L YG + S
Sbjct 126 ---------KFVRERTDKDIFGYPHSNNSCRLMDLLGYGKPITSS--------------- 161
Query 191 VATADSAVYSQKYNVNLAVNLFFLATYQKIYQDFFRWSQWE 231
T +Y+ VNLF L Y KIY D++R + +E
Sbjct 162 -KTPVPLLYTGN------VNLFRLLAYNKIYSDYYRNTTYE 195
Lambda K H a alpha
0.322 0.135 0.433 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 949922796600