bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-35_CDS_annotation_glimmer3.pl_2_6
Length=118
Score E
Sequences producing significant alignments: (Bits) Value
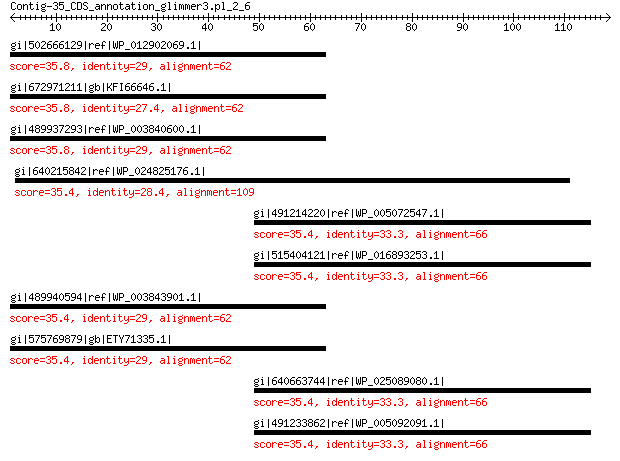
gi|502666129|ref|WP_012902069.1| sensory histidine kinase 35.8 4.1
gi|672971211|gb|KFI66646.1| histidine kinase-like protein 35.8 4.1
gi|489937293|ref|WP_003840600.1| sensory histidine kinase 35.8 4.3
gi|640215842|ref|WP_024825176.1| cyclase 35.4 4.7
gi|491214220|ref|WP_005072547.1| hypothetical protein 35.4 5.0
gi|515404121|ref|WP_016893253.1| hypothetical protein 35.4 5.0
gi|489940594|ref|WP_003843901.1| sensory histidine kinase 35.4 5.1
gi|575769879|gb|ETY71335.1| sensory histidine kinase 35.4 5.3
gi|640663744|ref|WP_025089080.1| MULTISPECIES: hypothetical protein 35.4 5.3
gi|491233862|ref|WP_005092091.1| hypothetical protein 35.4 5.3
>gi|502666129|ref|WP_012902069.1| sensory histidine kinase [Bifidobacterium dentium]
gi|283455794|ref|YP_003360358.1| histidine kinase-like protein [Bifidobacterium dentium Bd1]
gi|283102428|gb|ADB09534.1| Histidine kinase-like protein [Bifidobacterium dentium Bd1]
Length=508
Score = 35.8 bits (81), Expect = 4.1, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 33/64 (52%), Gaps = 2/64 (3%)
Query 1 VQNKFLTYKNDNIMKYSFPTKNN--DRLKSVEIYEGESIETKCARILQNKEPITDTAPII 58
V + F+ D I++Y+ P + RL SV +GE + R+L +P+ +T P++
Sbjct 187 VSDGFIVLTVDGIVRYASPNAISCFRRLGSVSTMQGEYLSEIGTRLLHENDPVLETLPLV 246
Query 59 YTAK 62
+ K
Sbjct 247 LSGK 250
>gi|672971211|gb|KFI66646.1| histidine kinase-like protein [Bifidobacterium kashiwanohense
JCM 15439 = DSM 21854]
Length=354
Score = 35.8 bits (81), Expect = 4.1, Method: Compositional matrix adjust.
Identities = 17/64 (27%), Positives = 34/64 (53%), Gaps = 2/64 (3%)
Query 1 VQNKFLTYKNDNIMKYSFPTKNN--DRLKSVEIYEGESIETKCARILQNKEPITDTAPII 58
V + F+ D I++Y+ P + RL SV +GE + ++L+ +P+ +T P++
Sbjct 187 VADGFIVLTVDGIVRYASPNAISCFRRLGSVSTMQGEYLSEIGTKLLRENDPVLETLPLV 246
Query 59 YTAK 62
+ K
Sbjct 247 LSGK 250
>gi|489937293|ref|WP_003840600.1| sensory histidine kinase [Bifidobacterium dentium]
gi|171278842|gb|EDT46503.1| ATPase/histidine kinase/DNA gyrase B/HSP90 domain protein [Bifidobacterium
dentium ATCC 27678]
gi|304553703|gb|EFM41614.1| ATPase/histidine kinase/DNA gyrase B/HSP90 domain protein [Bifidobacterium
dentium ATCC 27679]
Length=498
Score = 35.8 bits (81), Expect = 4.3, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 33/64 (52%), Gaps = 2/64 (3%)
Query 1 VQNKFLTYKNDNIMKYSFPTKNN--DRLKSVEIYEGESIETKCARILQNKEPITDTAPII 58
V + F+ D I++Y+ P + RL SV +GE + R+L +P+ +T P++
Sbjct 177 VSDGFIVLTVDGIVRYASPNAISCFRRLGSVSTMQGEYLSEIGTRLLHENDPVLETLPLV 236
Query 59 YTAK 62
+ K
Sbjct 237 LSGK 240
>gi|640215842|ref|WP_024825176.1| cyclase [Desulfovibrio magneticus]
Length=278
Score = 35.4 bits (80), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 31/119 (26%), Positives = 51/119 (43%), Gaps = 12/119 (10%)
Query 2 QNKFLTYKNDNIMKYSFPTK---NNDRLKSVEIYEGESIETKCA-----RILQNKEPITD 53
Q F+ + D ++ P K N K + Y G S+E A +I + TD
Sbjct 157 QGAFVAMRTDWSKRW--PDKAAMENKDAKGIAHYPGWSLEVLRALYEDRKITASGHETTD 214
Query 54 TAPIIYTAKEDGVLPAYNIRTDRFDIAMDAYDKITRSSAKKEIA--PKPEDFGNVPNKT 110
T P + T K+D L Y +++D + I + A+ + +A PKP+D P +
Sbjct 215 TDPGLTTTKDDYSLETYILKSDHYQIELLAHLDAVPEAGALVVASFPKPKDGSGFPARV 273
>gi|491214220|ref|WP_005072547.1| hypothetical protein [Mycobacterium abscessus]
gi|364001774|gb|EHM22966.1| hypothetical protein MBOL_03310 [Mycobacterium abscessus subsp.
bolletii BD]
Length=442
Score = 35.4 bits (80), Expect = 5.0, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 4/68 (6%)
Query 49 EPITDTAPIIYTAKEDGVLPAYNIRTDR--FDIAMDAYDKITRSSAKKEIAPKPEDFGNV 106
EP+ P + A +G L ++R R FD+ DA D TR +A+K++A +FG
Sbjct 100 EPLRPALPYVAHALREGRLGDEHVRIIRRFFDVLPDAVDADTREAAEKQLASMGTEFG-- 157
Query 107 PNKTEGGS 114
P + G+
Sbjct 158 PEQLRAGA 165
>gi|515404121|ref|WP_016893253.1| hypothetical protein [Mycobacterium abscessus]
Length=442
Score = 35.4 bits (80), Expect = 5.0, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 4/68 (6%)
Query 49 EPITDTAPIIYTAKEDGVLPAYNIRTDR--FDIAMDAYDKITRSSAKKEIAPKPEDFGNV 106
EP+ P + A +G L ++R R FD+ DA D TR +A+K++A +FG
Sbjct 100 EPLRPALPYVAHALREGRLGDEHVRIIRRFFDVLPDAVDADTREAAEKQLASMGTEFG-- 157
Query 107 PNKTEGGS 114
P + G+
Sbjct 158 PEQLRAGA 165
>gi|489940594|ref|WP_003843901.1| sensory histidine kinase [Bifidobacterium dentium]
gi|308222150|gb|EFO78433.1| ATPase/histidine kinase/DNA gyrase B/HSP90 domain protein [Bifidobacterium
dentium JCVIHMP022]
gi|570844056|gb|ETO97747.1| signal transduction histidine kinase [Bifidobacterium sp. MSTE12]
Length=494
Score = 35.4 bits (80), Expect = 5.1, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 33/64 (52%), Gaps = 2/64 (3%)
Query 1 VQNKFLTYKNDNIMKYSFPTKNN--DRLKSVEIYEGESIETKCARILQNKEPITDTAPII 58
V + F+ D I++Y+ P + RL SV +GE + R+L +P+ +T P++
Sbjct 173 VSDGFIVLTVDGIVRYASPNAISCFRRLGSVSTMQGEYLSEIGTRLLHENDPVLETLPLV 232
Query 59 YTAK 62
+ K
Sbjct 233 LSGK 236
>gi|575769879|gb|ETY71335.1| sensory histidine kinase [Bifidobacterium moukalabense DSM 27321]
Length=494
Score = 35.4 bits (80), Expect = 5.3, Method: Composition-based stats.
Identities = 18/64 (28%), Positives = 33/64 (52%), Gaps = 2/64 (3%)
Query 1 VQNKFLTYKNDNIMKYSFPTKNN--DRLKSVEIYEGESIETKCARILQNKEPITDTAPII 58
V + F+ D I++Y+ P + RL SV +GE + R+L +P+ +T P++
Sbjct 173 VSDGFIVLTVDGIVRYASPNAISCFRRLGSVSTMQGEYLSEIGTRLLHENDPVLETLPLV 232
Query 59 YTAK 62
+ K
Sbjct 233 LSGK 236
>gi|640663744|ref|WP_025089080.1| MULTISPECIES: hypothetical protein [Mycobacterium]
Length=442
Score = 35.4 bits (80), Expect = 5.3, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 4/68 (6%)
Query 49 EPITDTAPIIYTAKEDGVLPAYNIRTDR--FDIAMDAYDKITRSSAKKEIAPKPEDFGNV 106
EP+ P + A +G L ++R R FD+ DA D TR +A+K++A +FG
Sbjct 100 EPLRPALPYVAHALREGRLGDEHVRIIRRFFDVLPDAVDADTREAAEKQLASMGTEFG-- 157
Query 107 PNKTEGGS 114
P + G+
Sbjct 158 PEQLRAGA 165
>gi|491233862|ref|WP_005092091.1| hypothetical protein [Mycobacterium abscessus]
gi|169627498|ref|YP_001701147.1| Conserved hypothetical protein [Mycobacterium abscessus]
gi|169239465|emb|CAM60493.1| Conserved hypothetical protein [Mycobacterium abscessus]
gi|392067573|gb|EIT93421.1| hypothetical protein MA4S0726RB_4767 [Mycobacterium abscessus
4S-0726-RB]
gi|392075126|gb|EIU00960.1| hypothetical protein MA4S0726RA_0273 [Mycobacterium abscessus
4S-0726-RA]
gi|392077371|gb|EIU03202.1| hypothetical protein MA4S0303_0548 [Mycobacterium abscessus 4S-0303]
gi|392115025|gb|EIU40794.1| hypothetical protein MA6G0125R_4566 [Mycobacterium abscessus
6G-0125-R]
gi|392125722|gb|EIU51475.1| hypothetical protein MA6G0125S_0298 [Mycobacterium abscessus
6G-0125-S]
gi|392131146|gb|EIU56892.1| hypothetical protein MA6G0728S_1923 [Mycobacterium abscessus
6G-0728-S]
gi|392147528|gb|EIU73248.1| hypothetical protein MA6G1108_0296 [Mycobacterium abscessus 6G-1108]
gi|392175591|gb|EIV01253.1| hypothetical protein MA6G0212_0361 [Mycobacterium abscessus 6G-0212]
gi|392176644|gb|EIV02302.1| hypothetical protein MA6G0728R_0295 [Mycobacterium abscessus
6G-0728-R]
gi|392184849|gb|EIV10500.1| hypothetical protein MA4S0206_2001 [Mycobacterium abscessus 4S-0206]
gi|392204756|gb|EIV30341.1| hypothetical protein MA3A0119R_0336 [Mycobacterium abscessus
3A-0119-R]
gi|392214884|gb|EIV40433.1| hypothetical protein MA3A0122R_0402 [Mycobacterium abscessus
3A-0122-R]
gi|392217887|gb|EIV43420.1| hypothetical protein MA3A0122S_0156 [Mycobacterium abscessus
3A-0122-S]
gi|392218011|gb|EIV43543.1| hypothetical protein MA3A0731_0337 [Mycobacterium abscessus 3A-0731]
gi|392225979|gb|EIV51493.1| hypothetical protein MA4S0116R_0541 [Mycobacterium abscessus
4S-0116-R]
gi|392232798|gb|EIV58298.1| hypothetical protein MA3A0930S_0346 [Mycobacterium abscessus
3A-0930-S]
gi|392235898|gb|EIV61396.1| hypothetical protein MA4S0116S_4308 [Mycobacterium abscessus
4S-0116-S]
gi|392236492|gb|EIV61988.1| hypothetical protein MA3A0930R_0346 [Mycobacterium abscessus
3A-0930-R]
gi|392258887|gb|EIV84329.1| hypothetical protein MM3A0810R_0345 [Mycobacterium abscessus
3A-0810-R]
gi|528869702|gb|EPZ20995.1| hypothetical protein M879_08645 [Mycobacterium abscessus V06705]
gi|576369741|gb|ETZ63949.1| hypothetical protein L836_5398 [Mycobacterium abscessus MAB_110811_2726]
gi|576470578|gb|EUA64172.1| hypothetical protein I542_4339 [Mycobacterium abscessus 1948]
gi|576479485|gb|EUA73025.1| hypothetical protein I541_5234 [Mycobacterium chelonae 1518]
gi|576488182|gb|EUA81701.1| hypothetical protein I544_3010 [Mycobacterium abscessus 103]
Length=442
Score = 35.4 bits (80), Expect = 5.3, Method: Composition-based stats.
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 4/68 (6%)
Query 49 EPITDTAPIIYTAKEDGVLPAYNIRTDR--FDIAMDAYDKITRSSAKKEIAPKPEDFGNV 106
EP+ P + A +G L ++R R FD+ DA D TR +A+K++A +FG
Sbjct 100 EPLRPALPYVAHALREGRLGDEHVRIIRRFFDVLPDAVDADTREAAEKQLASMGTEFG-- 157
Query 107 PNKTEGGS 114
P + G+
Sbjct 158 PEQLRAGA 165
Lambda K H a alpha
0.310 0.130 0.366 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 442112487102