bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
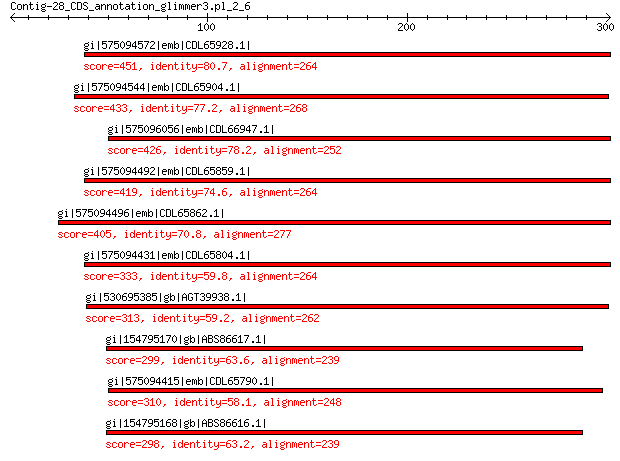
Query= Contig-28_CDS_annotation_glimmer3.pl_2_6
Length=301
Score E
Sequences producing significant alignments: (Bits) Value
gi|575094572|emb|CDL65928.1| unnamed protein product 451 2e-152
gi|575094544|emb|CDL65904.1| unnamed protein product 433 1e-145
gi|575096056|emb|CDL66947.1| unnamed protein product 426 1e-142
gi|575094492|emb|CDL65859.1| unnamed protein product 419 2e-140
gi|575094496|emb|CDL65862.1| unnamed protein product 405 2e-134
gi|575094431|emb|CDL65804.1| unnamed protein product 333 2e-106
gi|530695385|gb|AGT39938.1| major capsid protein 313 1e-99
gi|154795170|gb|ABS86617.1| capsid protein 299 9e-98
gi|575094415|emb|CDL65790.1| unnamed protein product 310 1e-97
gi|154795168|gb|ABS86616.1| capsid protein 298 2e-97
>gi|575094572|emb|CDL65928.1| unnamed protein product [uncultured bacterium]
Length=556
Score = 451 bits (1159), Expect = 2e-152, Method: Compositional matrix adjust.
Identities = 213/265 (80%), Positives = 234/265 (88%), Gaps = 2/265 (1%)
Query 38 NLWAVGDG-VATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQRPEY 96
N+WAV G V ATINQLRLAFQ+QKLYEKDARGGTRYTEI+RSHFGV SPDSRLQRPEY
Sbjct 293 NMWAVESGDVGMATINQLRLAFQLQKLYEKDARGGTRYTEIIRSHFGVVSPDSRLQRPEY 352
Query 97 LGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEHGFILGLMVARY 156
LGGNRIPI +NQI+QQS + E S P G G+S+T+D + DF KSF EHG+I+GL+VARY
Sbjct 353 LGGNRIPINVNQIIQQSQSTEQS-PLGALAGMSVTTDKNSDFIKSFVEHGYIIGLVVARY 411
Query 157 DHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQEAWADYR 216
DHTYQQGLDRM+SRK RFD+YWPV ANIGEQAVLNKEIY G++ DDEVFGYQEAWA+YR
Sbjct 412 DHTYQQGLDRMWSRKDRFDFYWPVLANIGEQAVLNKEIYIDGSDTDDEVFGYQEAWAEYR 471
Query 217 YKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSSVSNQLF 276
YKPNRVCGEMRS APQSLDVWHLGDDYS LP LSD WIREDKTNVDRVLAV SSVS+QLF
Sbjct 472 YKPNRVCGEMRSSAPQSLDVWHLGDDYSSLPYLSDSWIREDKTNVDRVLAVTSSVSDQLF 531
Query 277 ADIYVQNRCTRPMPMYSIPGLIDHH 301
ADIY+ N+ TRPMPMYSIPGLIDHH
Sbjct 532 ADIYICNKATRPMPMYSIPGLIDHH 556
>gi|575094544|emb|CDL65904.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 433 bits (1113), Expect = 1e-145, Method: Compositional matrix adjust.
Identities = 207/273 (76%), Positives = 229/273 (84%), Gaps = 6/273 (2%)
Query 33 LPVID-----NLWAVGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSP 87
+P ID NL A A+INQLRLAFQIQ+LYE+DARGGTRY EIL+SHFGVTSP
Sbjct 279 IPTIDAFRPLNLVANLQNATAASINQLRLAFQIQRLYERDARGGTRYIEILKSHFGVTSP 338
Query 88 DSRLQRPEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEHGF 147
D+RLQRPEYLGGNRIPI INQ++QQS T S PQGNPVG SLT+D + DF KSF EHGF
Sbjct 339 DARLQRPEYLGGNRIPININQVLQQSETTSTS-PQGNPVGQSLTTDTNADFVKSFVEHGF 397
Query 148 ILGLMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFG 207
++GLMVARYDHTYQQGL+R +SRK RFDYYWPVFA+IGEQAVLNKEIY GT DDEVFG
Sbjct 398 VIGLMVARYDHTYQQGLERFWSRKDRFDYYWPVFAHIGEQAVLNKEIYTSGTAVDDEVFG 457
Query 208 YQEAWADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAV 267
YQEA+ADYRYKP+RV GEMRS APQSLDVWHL DDY+ LPSLSD WIRE + VDRVLAV
Sbjct 458 YQEAYADYRYKPSRVTGEMRSAAPQSLDVWHLADDYASLPSLSDSWIRESASTVDRVLAV 517
Query 268 QSSVSNQLFADIYVQNRCTRPMPMYSIPGLIDH 300
S+VS QLF DIY+QNR TRPMPMYS+PGLIDH
Sbjct 518 SSNVSAQLFCDIYIQNRSTRPMPMYSVPGLIDH 550
>gi|575096056|emb|CDL66947.1| unnamed protein product [uncultured bacterium]
Length=570
Score = 426 bits (1094), Expect = 1e-142, Method: Compositional matrix adjust.
Identities = 197/253 (78%), Positives = 218/253 (86%), Gaps = 1/253 (0%)
Query 50 TINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQRPEYLGGNRIPIRINQI 109
TINQLR+AFQIQK YEK ARGG+RYTE++RS FGVTSPD+RLQR EYLGGNRIPI INQ+
Sbjct 318 TINQLRMAFQIQKFYEKQARGGSRYTEVIRSFFGVTSPDARLQRSEYLGGNRIPININQV 377
Query 110 VQQSATQEGST-PQGNPVGLSLTSDNHGDFTKSFTEHGFILGLMVARYDHTYQQGLDRMF 168
+QQS T ST PQG VG+S T+D H DFTKSFTEHGFI+G+M ARYDHTYQQG+DRM+
Sbjct 378 IQQSGTGSASTTPQGTVVGMSQTTDTHSDFTKSFTEHGFIIGVMCARYDHTYQQGIDRMW 437
Query 169 SRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQEAWADYRYKPNRVCGEMRS 228
SRK +FDYYWPVF+NIGEQA+ NKEIYAQG DDEVFGYQEAWA+YRYKP+RV GEMRS
Sbjct 438 SRKDKFDYYWPVFSNIGEQAIKNKEIYAQGNATDDEVFGYQEAWAEYRYKPSRVTGEMRS 497
Query 229 QAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSSVSNQLFADIYVQNRCTRP 288
QSLDVWHL DDYSKLPSLSDEWIRED ++RVLAV SNQ FADIYV+N CTRP
Sbjct 498 SYAQSLDVWHLADDYSKLPSLSDEWIREDAKTLNRVLAVSDQNSNQFFADIYVKNLCTRP 557
Query 289 MPMYSIPGLIDHH 301
MPMYSIPGLIDHH
Sbjct 558 MPMYSIPGLIDHH 570
>gi|575094492|emb|CDL65859.1| unnamed protein product [uncultured bacterium]
Length=551
Score = 419 bits (1077), Expect = 2e-140, Method: Compositional matrix adjust.
Identities = 197/267 (74%), Positives = 222/267 (83%), Gaps = 5/267 (2%)
Query 38 NLWA---VGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQRP 94
NLWA + ATINQLR AFQIQKLYE+DARGGTRY EIL+SHFGVTSPD+RLQRP
Sbjct 287 NLWADLSTATDLPVATINQLRTAFQIQKLYERDARGGTRYIEILKSHFGVTSPDARLQRP 346
Query 95 EYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEHGFILGLMVA 154
EYLGG+R+PI INQ++Q S T G+TPQGN SLT+D+H +FTKSF EHGFI+GLMVA
Sbjct 347 EYLGGSRVPININQVIQSSET--GATPQGNAAAYSLTTDSHSEFTKSFVEHGFIIGLMVA 404
Query 155 RYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQEAWAD 214
RYDH+YQQGL R +SRK RFDYYWPVFAN+GE AV NKEI+AQGT+ DDEVFGYQEAWAD
Sbjct 405 RYDHSYQQGLQRFWSRKDRFDYYWPVFANLGEMAVKNKEIFAQGTDVDDEVFGYQEAWAD 464
Query 215 YRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSSVSNQ 274
YRYKP+ V GEMRSQ QSLD+WHL DDY LPSLSD WIRED + V+RVLAV SVS Q
Sbjct 465 YRYKPSVVTGEMRSQYAQSLDIWHLADDYENLPSLSDSWIREDSSTVNRVLAVSDSVSAQ 524
Query 275 LFADIYVQNRCTRPMPMYSIPGLIDHH 301
LF DIY++ TRPMP+YSIPGLIDHH
Sbjct 525 LFCDIYIRCLATRPMPLYSIPGLIDHH 551
>gi|575094496|emb|CDL65862.1| unnamed protein product [uncultured bacterium]
Length=568
Score = 405 bits (1040), Expect = 2e-134, Method: Compositional matrix adjust.
Identities = 196/277 (71%), Positives = 225/277 (81%), Gaps = 5/277 (2%)
Query 25 SAGSSEDALPVIDNLWAVGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGV 84
S GS P DNL+A G AT TINQLR+AFQIQKLYEKDAR G+RY E++RSHF V
Sbjct 297 SFGSKLSVYP--DNLYA-SSGTAT-TINQLRMAFQIQKLYEKDARAGSRYRELIRSHFSV 352
Query 85 TSPDSRLQRPEYLGGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTE 144
T D+R+Q PEYLGGNRIPI INQ+VQ S T + S PQGN G SLTSD+HGDF KSFTE
Sbjct 353 TPLDARMQVPEYLGGNRIPININQVVQTSQTSDVS-PQGNVAGQSLTSDSHGDFIKSFTE 411
Query 145 HGFILGLMVARYDHTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDE 204
HG ++G+ VARYDHTYQQG+ +++SRK+RFDYYWPV ANIGEQAVLNKEIYAQGT +D+E
Sbjct 412 HGMLIGVAVARYDHTYQQGVSKLWSRKTRFDYYWPVLANIGEQAVLNKEIYAQGTAQDEE 471
Query 205 VFGYQEAWADYRYKPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRV 264
VFGYQEAWA+YRYKP+ V GEMRS A SLD WH DDY+ LP LS +WI+EDKTN+DRV
Sbjct 472 VFGYQEAWAEYRYKPSIVTGEMRSSARTSLDSWHFADDYNSLPKLSADWIKEDKTNIDRV 531
Query 265 LAVQSSVSNQLFADIYVQNRCTRPMPMYSIPGLIDHH 301
LAV SSVSNQ FAD Y++N TR +P YSIPGLIDHH
Sbjct 532 LAVSSSVSNQYFADFYIENETTRALPFYSIPGLIDHH 568
>gi|575094431|emb|CDL65804.1| unnamed protein product [uncultured bacterium]
Length=560
Score = 333 bits (853), Expect = 2e-106, Method: Compositional matrix adjust.
Identities = 158/264 (60%), Positives = 195/264 (74%), Gaps = 4/264 (2%)
Query 38 NLWAVGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQRPEYL 97
NLWA A AT+NQLR AFQ+QKL EKDARGGTRY EIL++HFGVT+ D+R+Q PEYL
Sbjct 301 NLWA-SPVTAAATVNQLRQAFQVQKLLEKDARGGTRYREILKNHFGVTTSDARMQIPEYL 359
Query 98 GGNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEHGFILGLMVARYD 157
GG ++PI ++Q+VQ SA+ + S PQGN +S+T + FTKSF EHGFI+G+ AR
Sbjct 360 GGCKVPINVSQVVQTSASTDAS-PQGNTAAISVTPFSKSMFTKSFDEHGFIIGVATARTA 418
Query 158 HTYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQEAWADYRY 217
+YQQG++RM+SRK R DYY+PV ANIGEQA+LNKEIYAQG +DDE FGYQEAWADYRY
Sbjct 419 QSYQQGIERMWSRKDRLDYYFPVLANIGEQAILNKEIYAQGNAKDDEAFGYQEAWADYRY 478
Query 218 KPNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSSVSNQLFA 277
KPN +CG RS A QSLD WH G DY KLP+LS +W+ + + R LAVQ+ A
Sbjct 479 KPNTICGRFRSNAQQSLDAWHYGQDYDKLPTLSTDWMEQSDIEMKRTLAVQTEP--DFIA 536
Query 278 DIYVQNRCTRPMPMYSIPGLIDHH 301
+ + R MP+YSIPGLIDH+
Sbjct 537 NFRFNCKTVRVMPLYSIPGLIDHN 560
>gi|530695385|gb|AGT39938.1| major capsid protein [Marine gokushovirus]
Length=514
Score = 313 bits (803), Expect = 1e-99, Method: Compositional matrix adjust.
Identities = 155/262 (59%), Positives = 190/262 (73%), Gaps = 4/262 (2%)
Query 39 LWAVGDGVATATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQRPEYLG 98
+WA ATINQLR AFQIQ+LYEKDARGGTRYTE+++SHFGVTSPD+RLQRPEYLG
Sbjct 256 IWADLSDATAATINQLREAFQIQRLYEKDARGGTRYTEVIQSHFGVTSPDARLQRPEYLG 315
Query 99 GNRIPIRINQIVQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEHGFILGLMVARYDH 158
G + I IN I Q S+T + +TPQGN G T F KSFTEH +LGL D
Sbjct 316 GGKDRININPIAQTSST-DATTPQGNLSGYGTTGFTGHRFNKSFTEHSVVLGLACVFADL 374
Query 159 TYQQGLDRMFSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQEAWADYRYK 218
TYQQGL R FSR++R+D+YWP A++GEQAVLNKEIYAQGT +D+ VFGYQE +A+YRYK
Sbjct 375 TYQQGLPRHFSRQTRWDFYWPALAHLGEQAVLNKEIYAQGTTDDNNVFGYQERYAEYRYK 434
Query 219 PNRVCGEMRSQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSSVSNQLFAD 278
P+ + G+MRS QSLD+WHL D+ LP L+ +I E+ VDRV AVQ+ + L D
Sbjct 435 PSSITGQMRSNFAQSLDIWHLAQDFGSLPVLNSSFIEENPP-VDRVTAVQNYPN--LILD 491
Query 279 IYVQNRCTRPMPMYSIPGLIDH 300
+Y + +C RPMP Y +PGLIDH
Sbjct 492 MYFKLKCARPMPTYGVPGLIDH 513
>gi|154795170|gb|ABS86617.1| capsid protein [uncultured phage]
Length=234
Score = 299 bits (766), Expect = 9e-98, Method: Compositional matrix adjust.
Identities = 152/240 (63%), Positives = 180/240 (75%), Gaps = 7/240 (3%)
Query 49 ATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQRPEYLGGNRIPIRINQ 108
ATINQLR +FQIQKLYE+DARGGTRYTEI+RSHFGVTSPD+RLQRPEYLGG PI +N
Sbjct 1 ATINQLRQSFQIQKLYERDARGGTRYTEIIRSHFGVTSPDARLQRPEYLGGGSTPINVNP 60
Query 109 IVQQSATQEGSTPQGNPVGLSLT-SDNHGDFTKSFTEHGFILGLMVARYDHTYQQGLDRM 167
I Q + G+TPQGN + D HG FTKSFTEH ++G++ AR D TYQQGL+RM
Sbjct 61 IAQTG--ESGTTPQGNLAAMGTAYMDGHG-FTKSFTEHCVVIGIVSARADLTYQQGLNRM 117
Query 168 FSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQEAWADYRYKPNRVCGEMR 227
+SR +R+D+YWP A+IGEQAVLNKEIYAQGT+ DDEVFGYQE +A+YRYKP+ G MR
Sbjct 118 WSRSTRWDFYWPALAHIGEQAVLNKEIYAQGTSADDEVFGYQERFAEYRYKPSLTTGLMR 177
Query 228 SQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSSVSNQLFADIYVQNRCTR 287
S A SLD WHLG D+S LP+L+ +I+ED VDRV+AV S D Y Q RC R
Sbjct 178 SNATTSLDTWHLGVDFSTLPALNAAFIQEDPP-VDRVIAVPS--EPHFLFDSYFQYRCAR 234
>gi|575094415|emb|CDL65790.1| unnamed protein product [uncultured bacterium]
Length=569
Score = 310 bits (793), Expect = 1e-97, Method: Compositional matrix adjust.
Identities = 144/248 (58%), Positives = 182/248 (73%), Gaps = 1/248 (0%)
Query 50 TINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQRPEYLGGNRIPIRINQI 109
+IN LR A +Q + E DARGGTRY EIL++ FGV+SPD+RLQR EY+GG RIPI ++Q+
Sbjct 320 SINDLRQAIALQHILEADARGGTRYVEILKNEFGVSSPDARLQRSEYIGGERIPINVSQV 379
Query 110 VQQSATQEGSTPQGNPVGLSLTSDNHGDFTKSFTEHGFILGLMVARYDHTYQQGLDRMFS 169
+Q SA+ + ++PQGN SLT+ + S EHG+ILGL R DH+YQQGL RM++
Sbjct 380 IQSSAS-DTTSPQGNAAAYSLTTSANTIRAYSAVEHGYILGLAAIRVDHSYQQGLSRMWT 438
Query 170 RKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQEAWADYRYKPNRVCGEMRSQ 229
R RF YY P+ AN+GEQAVLN+EIYAQGT D EVFGYQEAWADYRY+ N + GEMRS
Sbjct 439 RSDRFSYYHPMLANLGEQAVLNQEIYAQGTTADTEVFGYQEAWADYRYRTNMITGEMRST 498
Query 230 APQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSSVSNQLFADIYVQNRCTRPM 289
QSLD WH GD Y+ LP LS++WI+E + N+DR LAVQS S+Q ++Y RPM
Sbjct 499 YAQSLDAWHYGDKYTDLPRLSNDWIKEGQENIDRTLAVQSENSHQFICNLYFDQTWVRPM 558
Query 290 PMYSIPGL 297
P+YS+PGL
Sbjct 559 PIYSVPGL 566
>gi|154795168|gb|ABS86616.1| capsid protein [uncultured phage]
gi|154795177|gb|ABS86620.1| capsid protein [uncultured phage]
Length=234
Score = 298 bits (763), Expect = 2e-97, Method: Compositional matrix adjust.
Identities = 151/240 (63%), Positives = 180/240 (75%), Gaps = 7/240 (3%)
Query 49 ATINQLRLAFQIQKLYEKDARGGTRYTEILRSHFGVTSPDSRLQRPEYLGGNRIPIRINQ 108
ATINQLR +FQIQKLYE+DARGGTRYTEI+RSHFGVTSPD+RLQRPEYLGG PI +N
Sbjct 1 ATINQLRQSFQIQKLYERDARGGTRYTEIIRSHFGVTSPDARLQRPEYLGGGSTPINVNP 60
Query 109 IVQQSATQEGSTPQGNPVGLSLT-SDNHGDFTKSFTEHGFILGLMVARYDHTYQQGLDRM 167
I Q + G+TPQGN + D HG FTKSFTEH ++G++ AR D TYQQGL+RM
Sbjct 61 IAQTG--ESGTTPQGNLAAMGTAYMDGHG-FTKSFTEHCVVIGIVSARADLTYQQGLNRM 117
Query 168 FSRKSRFDYYWPVFANIGEQAVLNKEIYAQGTNEDDEVFGYQEAWADYRYKPNRVCGEMR 227
+SR +R+D+YWP A+IGEQAVLNKEIYAQGT+ DD+VFGYQE +A+YRYKP+ G MR
Sbjct 118 WSRSTRWDFYWPALAHIGEQAVLNKEIYAQGTSADDDVFGYQERFAEYRYKPSLTTGLMR 177
Query 228 SQAPQSLDVWHLGDDYSKLPSLSDEWIREDKTNVDRVLAVQSSVSNQLFADIYVQNRCTR 287
S A SLD WHLG D+S LP+L+ +I+ED VDRV+AV S D Y Q RC R
Sbjct 178 SNATTSLDTWHLGQDFSALPALNAAFIQEDPP-VDRVIAVPS--EPHFLFDSYFQYRCAR 234
Lambda K H a alpha
0.317 0.134 0.404 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1595232544752