bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-26_CDS_annotation_glimmer3.pl_2_1
Length=295
Score E
Sequences producing significant alignments: (Bits) Value
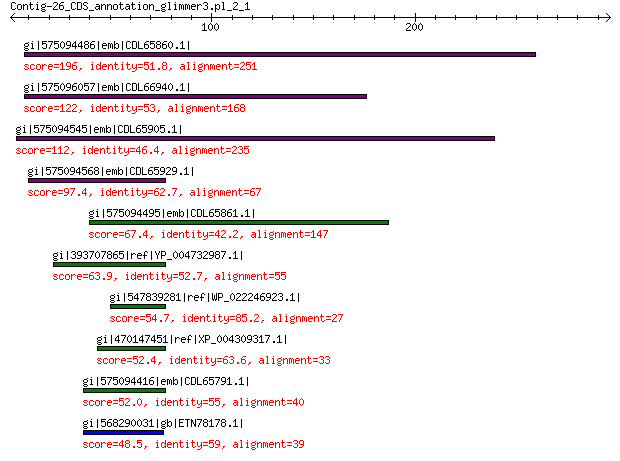
gi|575094486|emb|CDL65860.1| unnamed protein product 196 2e-56
gi|575096057|emb|CDL66940.1| unnamed protein product 122 6e-29
gi|575094545|emb|CDL65905.1| unnamed protein product 112 2e-25
gi|575094568|emb|CDL65929.1| unnamed protein product 97.4 4e-20
gi|575094495|emb|CDL65861.1| unnamed protein product 67.4 9e-10
gi|393707865|ref|YP_004732987.1| structural protein VP2 63.9 8e-09
gi|547839281|ref|WP_022246923.1| putative minor capsid protein 54.7 1e-05
gi|470147451|ref|XP_004309317.1| PREDICTED: minor spike protein ... 52.4 3e-05
gi|575094416|emb|CDL65791.1| unnamed protein product 52.0 2e-04
gi|568290031|gb|ETN78178.1| hypothetical protein NECAME_18237 48.5 5e-04
>gi|575094486|emb|CDL65860.1| unnamed protein product [uncultured bacterium]
Length=344
Score = 196 bits (498), Expect = 2e-56, Method: Compositional matrix adjust.
Identities = 130/252 (52%), Positives = 179/252 (71%), Gaps = 12/252 (5%)
Query 8 LQGIAGQNTSSSAKQAEELRTWQEQQAELARKYNSQEAQKNRDWQERMSSTAHQREVRDL 67
LQ I N + SA QA++ +Q Q L R++N EA+ +R WQERMS+TAHQRE++DL
Sbjct 68 LQSITASNNAWSAAQAQKQMDFQASQGALVRQFNHDEAELSRLWQERMSNTAHQREIKDL 127
Query 68 IAAGLNPVLsvtggsgaavtsgatass-sapsgamgsvDNSATGAVAglfgsllssflsl 126
AAGLNPVLS GGSGA VTSG+TAS S PSG+ G D S GA+ L GS + + S+
Sbjct 128 QAAGLNPVLSAMGGSGAPVTSGSTASGYSPPSGSKGDTDTSLAGALVSLLGSSMMAQASM 187
Query 127 EGTRVSAQSNQAIADKYTAMSKYTSELQAQTQLTSTNIQAMAQKYTADAHLAGTKYAADQ 186
T +SA++ +++ADKYTAMSK +E+Q +T L+++ I AMA ++YAAD+
Sbjct 188 ANTAMSARTQESVADKYTAMSKLVAEIQQETTLSASTISAMA-----------SRYAADR 236
Query 187 SAAAQKVSASIHAAAQKYGYNVQSMTQRDIAAFNAQVNKDLQKAGFKQEFDIKKAFPNNA 246
SA A KV+ASIHAAAQ+YGY+VQ+MTQRDIA+FNAQVNKDL + G++ +FDIK+A+P++
Sbjct 237 SADASKVAASIHAAAQRYGYDVQAMTQRDIASFNAQVNKDLAQMGYQHDFDIKEAYPSSM 296
Query 247 WNVFGGLGTQAV 258
+ L +++
Sbjct 297 AGLMASLFGESI 308
>gi|575096057|emb|CDL66940.1| unnamed protein product [uncultured bacterium]
Length=275
Score = 122 bits (305), Expect = 6e-29, Method: Compositional matrix adjust.
Identities = 89/168 (53%), Positives = 118/168 (70%), Gaps = 0/168 (0%)
Query 8 LQGIAGQNTSSSAKQAEELRTWQEQQAELARKYNSQEAQKNRDWQERMSSTAHQREVRDL 67
++GIA N++ +A+QAE R WQE Q A ++NS EA KNR WQE MS+TAHQREV+DL
Sbjct 28 MKGIAQANSAWNAEQAEIQRDWQEAQNAKAMQFNSMEAAKNRKWQEMMSNTAHQREVKDL 87
Query 68 IAAGLNPVLsvtggsgaavtsgatasssapsgamgsvDNSATGAVAglfgsllssflslE 127
+AAGLNPVLS G+GAAV SGATAS +GA G D S +GA+A L GS+LS+ +++
Sbjct 88 MAAGLNPVLSAMNGNGAAVGSGATASGVTSAGAKGEADTSTSGAIANLLGSILSASTAIQ 147
Query 128 GTRVSAQSNQAIADKYTAMSKYTSELQAQTQLTSTNIQAMAQKYTADA 175
V+A++ +A+ADKYTAMS+ +E+ L S I A A +Y ADA
Sbjct 148 AANVNARTQEAVADKYTAMSQIVAEINKAATLGSAGIHAGATRYAADA 195
>gi|575094545|emb|CDL65905.1| unnamed protein product [uncultured bacterium]
Length=325
Score = 112 bits (281), Expect = 2e-25, Method: Compositional matrix adjust.
Identities = 109/246 (44%), Positives = 151/246 (61%), Gaps = 11/246 (4%)
Query 4 WAGALQGIAGQNTSSSAKQAEELRTWQEQQAELARKYNSQEAQKNRDWQERMSSTAHQRE 63
+ G + +A N++ +A QA R WQ+QQ +A +++S EA KNRDWQ MS+TAHQRE
Sbjct 23 YLGQITRMASDNSAFNASQAAANRNWQQQQNNIAMQFSSAEAAKNRDWQSYMSNTAHQRE 82
Query 64 VRDLIAAGLNPVLsvtggsgaavtsgatasssapsgamgsvDNSATGAVAglfgsllssf 123
V DL AAGLNPVLS GG+GAAVTSGATA SG S D SAT A+ GL GSLL++
Sbjct 83 VADLKAAGLNPVLSAMGGNGAAVTSGATAQGYTSSGGQASADTSATAALVGLLGSLLNAQ 142
Query 124 lslEGTRVSAQSNQAIADKYTAMSKYTSELQAQTQLTSTNIQAM-----------AQKYT 172
S+ T +A +N ++ADKYT+ ++Y +++ S N+ A A KY
Sbjct 143 TSIANTATNAVANLSVADKYTSATRYAADVGYAGTSYSANVAAYASRFASNNALAASKYA 202
Query 173 ADAHLAGTKYAADQSAAAQKVSASIHAAAQKYGYNVQSMTQRDIAAFNAQVNKDLQKAGF 232
+D A +KYA+DQS A K ++ + + KY + ++ T RD+A FNA VN+DLQK
Sbjct 203 SDNSRAASKYASDQSYLASKFASILQSNTAKYNIDTRTATDRDLAEFNAAVNRDLQKNEI 262
Query 233 KQEFDI 238
+F +
Sbjct 263 DAKFSL 268
>gi|575094568|emb|CDL65929.1| unnamed protein product [uncultured bacterium]
Length=310
Score = 97.4 bits (241), Expect = 4e-20, Method: Compositional matrix adjust.
Identities = 42/67 (63%), Positives = 57/67 (85%), Gaps = 0/67 (0%)
Query 10 GIAGQNTSSSAKQAEELRTWQEQQAELARKYNSQEAQKNRDWQERMSSTAHQREVRDLIA 69
G++ +NT+ S ++AE LRTWQE+Q +A ++N+ EA+KNR+WQE MS+TAHQREV DL+A
Sbjct 48 GLSEKNTARSVQEAESLRTWQEEQNRIAMQFNAAEAEKNRNWQEIMSNTAHQREVNDLMA 107
Query 70 AGLNPVL 76
AGLNPVL
Sbjct 108 AGLNPVL 114
>gi|575094495|emb|CDL65861.1| unnamed protein product [uncultured bacterium]
Length=266
Score = 67.4 bits (163), Expect = 9e-10, Method: Compositional matrix adjust.
Identities = 62/151 (41%), Positives = 82/151 (54%), Gaps = 18/151 (12%)
Query 40 YNSQEAQKNRDWQERMSSTAHQREVRDLIAAGLNPVLsvtggsgaavtsgatasssapsg 99
YN+Q A++ +QERMSSTAHQREV+DLIAAGLNPVL S + +SAPSG
Sbjct 66 YNTQSAREQMAFQERMSSTAHQREVKDLIAAGLNPVL-----------SAGGSGASAPSG 114
Query 100 amgsvDNSATGAVAglfgsllssflslEGTRVSAQSNQAIADKYTAMSKYTSELQAQTQL 159
AM + D+S A A L+ + N+A D AM+KY+ ++ AQT L
Sbjct 115 AMATADSSMMSAKANAALQKRIVNAQLKNAK---DINKAQLDAQKAMNKYSVDVGAQTSL 171
Query 160 TSTNIQAMAQKY----TADAHLAGTKYAADQ 186
+ I A A K+ A A + G+ AA Q
Sbjct 172 ANAQISASASKFGAMQAAAASMYGSNLAAKQ 202
>gi|393707865|ref|YP_004732987.1| structural protein VP2 [Microviridae phi-CA82]
gi|311336637|gb|ADP89808.1| structural protein VP2 [Microviridae phi-CA82]
Length=234
Score = 63.9 bits (154), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 29/55 (53%), Positives = 42/55 (76%), Gaps = 0/55 (0%)
Query 22 QAEELRTWQEQQAELARKYNSQEAQKNRDWQERMSSTAHQREVRDLIAAGLNPVL 76
QA++ W +Q E + ++N+QEAQKNRDWQE+MS+TA QR+++D AGLNP+
Sbjct 11 QADKQNKWNAEQTEKSNQFNAQEAQKNRDWQEQMSNTALQRKMQDAEKAGLNPIF 65
>gi|547839281|ref|WP_022246923.1| putative minor capsid protein [Clostridium sp. CAG:306]
gi|524476581|emb|CDC18646.1| putative minor capsid protein [Clostridium sp. CAG:306]
Length=236
Score = 54.7 bits (130), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 23/27 (85%), Positives = 26/27 (96%), Gaps = 0/27 (0%)
Query 50 DWQERMSSTAHQREVRDLIAAGLNPVL 76
D+QERMSSTAHQREV+DL AAGLNP+L
Sbjct 40 DFQERMSSTAHQREVKDLRAAGLNPIL 66
>gi|470147451|ref|XP_004309317.1| PREDICTED: minor spike protein H-like, partial [Fragaria vesca
subsp. vesca]
Length=139
Score = 52.4 bits (124), Expect = 3e-05, Method: Compositional matrix adjust.
Identities = 21/33 (64%), Positives = 31/33 (94%), Gaps = 0/33 (0%)
Query 44 EAQKNRDWQERMSSTAHQREVRDLIAAGLNPVL 76
EAQ+NR++QER+S++A+QR+V DL +AGLNP+L
Sbjct 25 EAQRNREFQERLSNSAYQRQVADLSSAGLNPML 57
>gi|575094416|emb|CDL65791.1| unnamed protein product [uncultured bacterium]
Length=311
Score = 52.0 bits (123), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 22/40 (55%), Positives = 29/40 (73%), Gaps = 0/40 (0%)
Query 37 ARKYNSQEAQKNRDWQERMSSTAHQREVRDLIAAGLNPVL 76
A++YN +EAQ R W + M TA+Q V+DL AAGLNP+L
Sbjct 144 AQRYNREEAQAERSWAQSMRQTAYQDTVKDLKAAGLNPIL 183
>gi|568290031|gb|ETN78178.1| hypothetical protein NECAME_18237 [Necator americanus]
Length=112
Score = 48.5 bits (114), Expect = 5e-04, Method: Compositional matrix adjust.
Identities = 23/39 (59%), Positives = 28/39 (72%), Gaps = 0/39 (0%)
Query 37 ARKYNSQEAQKNRDWQERMSSTAHQREVRDLIAAGLNPV 75
A K N + ++ WQERMS+TAHQRE DL AAGLNP+
Sbjct 30 ANKANRKMMREQMAWQERMSNTAHQREQADLKAAGLNPI 68
Lambda K H a alpha
0.310 0.120 0.334 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1550741951049