bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_6
Length=163
Score E
Sequences producing significant alignments: (Bits) Value
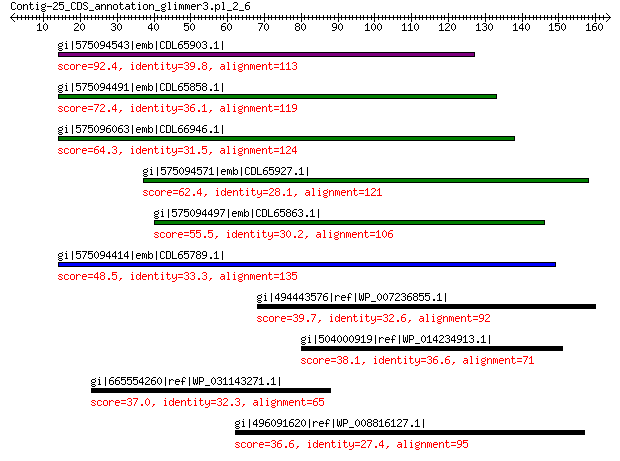
gi|575094543|emb|CDL65903.1| unnamed protein product 92.4 2e-20
gi|575094491|emb|CDL65858.1| unnamed protein product 72.4 6e-13
gi|575096063|emb|CDL66946.1| unnamed protein product 64.3 4e-10
gi|575094571|emb|CDL65927.1| unnamed protein product 62.4 1e-09
gi|575094497|emb|CDL65863.1| unnamed protein product 55.5 5e-07
gi|575094414|emb|CDL65789.1| unnamed protein product 48.5 2e-04
gi|494443576|ref|WP_007236855.1| DNA helicase 39.7 0.55
gi|504000919|ref|WP_014234913.1| chemotaxis protein 38.1 1.8
gi|665554260|ref|WP_031143271.1| G-D-S-L family lipolytic protein 37.0 3.5
gi|496091620|ref|WP_008816127.1| hypothetical protein 36.6 5.7
>gi|575094543|emb|CDL65903.1| unnamed protein product [uncultured bacterium]
Length=145
Score = 92.4 bits (228), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 45/114 (39%), Positives = 71/114 (62%), Gaps = 1/114 (1%)
Query 14 IYTASGSPIHRILEPRFDGVNTC-LVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSV 72
+Y +SGSPIH + + R+D L +G++NI D++++ S DI+ +L R GD V
Sbjct 12 VYCSSGSPIHIVYQGRYDDDGVFDLFPSGQDNIYDQIQSHRDSVDIHVLLDRYQRGDVDV 71
Query 73 LSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADL 126
LS+++ +YGDFTG+P+ E LN V E AF L D++AKY + + +W + L
Sbjct 72 LSARQGVYGDFTGMPASYSEILNAVLAGERAFMDLPVDERAKYGHSFAQWLSSL 125
>gi|575094491|emb|CDL65858.1| unnamed protein product [uncultured bacterium]
Length=176
Score = 72.4 bits (176), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 43/120 (36%), Positives = 62/120 (52%), Gaps = 5/120 (4%)
Query 14 IYTASGSPIHRILEPRFDGVNTC-LVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSV 72
++ G PI + PR+D L TGEEN+ D +++ A STDI+ +L R + G+T V
Sbjct 14 VFQRPGDPIKVVYSPRYDENGVLDLQPTGEENLYDFIQSHAQSTDIHVILDRFASGETDV 73
Query 73 LSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDN 132
LS + Y D + +P E LN V E F +L + K K+ N + W LS DN
Sbjct 74 LSQIQGFYADASDMPKTYAEVLNAVIAGEQTFDRLPVEIKQKFGNSFSTW----LSSMDN 129
>gi|575096063|emb|CDL66946.1| unnamed protein product [uncultured bacterium]
Length=163
Score = 64.3 bits (155), Expect = 4e-10, Method: Compositional matrix adjust.
Identities = 39/125 (31%), Positives = 62/125 (50%), Gaps = 3/125 (2%)
Query 14 IYTASGSPIHRILEPRFDGVN-TCLVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSV 72
I+T +GS H +D L TG N+ D +++ A S D++ ++ R S GD
Sbjct 14 IFTEAGSREHITYGGHYDDEGRVVLEETGRINLYDEIQSHAESVDLHVLMQRYSCGDVDC 73
Query 73 LSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDN 132
LS ++ +GD P EALN + + E F L D +AK+ + + + A SG D+
Sbjct 74 LSQRQGFFGDVLDFPQTYAEALNHMQEMERQFMSLPLDVRAKFGHSFSEFLA--ASGDDD 131
Query 133 LSEKL 137
E+L
Sbjct 132 FLERL 136
>gi|575094571|emb|CDL65927.1| unnamed protein product [uncultured bacterium]
Length=133
Score = 62.4 bits (150), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 34/121 (28%), Positives = 64/121 (53%), Gaps = 5/121 (4%)
Query 37 LVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSVLSSKRPMYGDFTGLPSDPIEALNL 96
LV G++N+ D +++ S D+N +L R + G+ VLS + Y D + +P + LNL
Sbjct 12 LVENGKKNLYDEIQSHKDSVDLNLLLQRFNNGEVDVLSRMQGTYADLSNMPKTYADMLNL 71
Query 97 VHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDNLSEKLSSVVPNSDVEKEGADSVVE 156
+ + E F L D +AK+++ + +W S + + ++ +S V++E D + E
Sbjct 72 IKKGEADFLSLPVDVRAKFDHSFEKWLVTFGS-----QDWIVNMKKDSVVQEEKTDILAE 126
Query 157 K 157
K
Sbjct 127 K 127
>gi|575094497|emb|CDL65863.1| unnamed protein product [uncultured bacterium]
Length=170
Score = 55.5 bits (132), Expect = 5e-07, Method: Compositional matrix adjust.
Identities = 32/106 (30%), Positives = 56/106 (53%), Gaps = 3/106 (3%)
Query 40 TGEENIQDRMEAEAPSTDINYMLHRLSLGDTSVLSSKRPMYGDFTGLPSDPIEALNLVHQ 99
G ++ ++++ S DINY+L R + GD S LS + +YGDFT +P++ E V
Sbjct 41 VGRIDLYAQIQSYKDSCDINYILERFARGDESALSKIQGVYGDFTAMPTNLAELQQRVVD 100
Query 100 SEYAFAQLSADDKAKYNNDWRRWFADLLSGRDNLSEKLSSVVPNSD 145
+E F L D +A++N+ +++ + G D + K + P D
Sbjct 101 AEALFYNLPVDIRAEFNHSPSEFYSAI--GTDKFN-KAVGITPQVD 143
>gi|575094414|emb|CDL65789.1| unnamed protein product [uncultured bacterium]
Length=167
Score = 48.5 bits (114), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 45/147 (31%), Positives = 69/147 (47%), Gaps = 16/147 (11%)
Query 14 IYTASGSPIHRI-------LEPRF-----DGVNTCLVVTGEENIQDRMEAEAPSTDINYM 61
IYTA G I LEP + + T L GE N +++ D++ +
Sbjct 5 IYTAYGPKREVIPNEPGNELEPHYIEKIGENGRTYLEKDGETNTYAEIQSYKDECDVHSI 64
Query 62 LHRLSLGDTSVLSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRR 121
L R GDTSVLS ++ +Y D T LP+ E NL+ + F QL + K K++N +
Sbjct 65 LCRYFAGDTSVLS-RQGVYIDATQLPTTYHEMYNLMAEQRDKFDQLPLEIKRKFDNSFNV 123
Query 122 WFADLLSGRDNLSEKLSSVVPNSDVEK 148
W + +G + KL +V N++ K
Sbjct 124 WAS--TAGSEEWY-KLMGIVQNAENNK 147
>gi|494443576|ref|WP_007236855.1| DNA helicase [Gordonia otitidis]
gi|377525044|dbj|GAB32589.1| hypothetical protein GOOTI_015_00040 [Gordonia otitidis NBRC
100426]
Length=1385
Score = 39.7 bits (91), Expect = 0.55, Method: Compositional matrix adjust.
Identities = 30/113 (27%), Positives = 49/113 (43%), Gaps = 21/113 (19%)
Query 68 GDTSVLSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQL-----SADDKAKYNNDWRR- 121
G + + +RP + GL +D + L+ VHQ + A L ++ ++K N R
Sbjct 632 GVIATVDGRRPTDAESLGLVADHLAVLDAVHQVQRVLADLHVPVDTSGSRSKQLNQLVRL 691
Query 122 -----WFADLLSGRDNLSEKLSSVVPN----------SDVEKEGADSVVEKDS 159
W +DLLS RD L +L + P ++V +EGA D+
Sbjct 692 DTQLSWVSDLLSARDQLIHELEYISPGGPRPRSVAEVAEVAREGASIAAANDA 744
>gi|504000919|ref|WP_014234913.1| chemotaxis protein [Vibrio sp. EJY3]
gi|375263822|ref|YP_005026052.1| methyl-accepting chemotaxis protein [Vibrio sp. EJY3]
gi|369844249|gb|AEX25077.1| methyl-accepting chemotaxis protein [Vibrio sp. EJY3]
Length=547
Score = 38.1 bits (87), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 26/71 (37%), Positives = 39/71 (55%), Gaps = 4/71 (6%)
Query 80 YGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRRWFADLLSGRDNLSEKLSS 139
Y DFT L P+ L H S+Y A+ + + +A N +R ++++ DNLSEKL+
Sbjct 122 YVDFTRLTMTPL--LTQKHASQYTTAEFNQNFEAAMVN-YRVAGEEMINAIDNLSEKLNQ 178
Query 140 VVPNSDVEKEG 150
V SDV+ G
Sbjct 179 TVI-SDVQANG 188
>gi|665554260|ref|WP_031143271.1| G-D-S-L family lipolytic protein [Streptomyces xanthophaeus]
Length=235
Score = 37.0 bits (84), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 0/65 (0%)
Query 23 HRILEPRFDGVNTCLVVTGEENIQDRMEAEAPSTDINYMLHRLSLGDTSVLSSKRPMYGD 82
HR+L DG + +V+ G ++ R AE S D+ ++ L+ TSV+ P +G+
Sbjct 93 HRLLPQLGDGFDLAVVLAGSNDVLGRRTAEEWSDDLAAIVDDLADRATSVVVVGTPPFGE 152
Query 83 FTGLP 87
F LP
Sbjct 153 FPSLP 157
>gi|496091620|ref|WP_008816127.1| hypothetical protein [Erysipelotrichaceae bacterium 6_1_45]
gi|371667179|gb|EHO32308.1| hypothetical protein HMPREF0981_00230 [Erysipelotrichaceae bacterium
6_1_45]
Length=306
Score = 36.6 bits (83), Expect = 5.7, Method: Compositional matrix adjust.
Identities = 26/101 (26%), Positives = 49/101 (49%), Gaps = 6/101 (6%)
Query 62 LHRLSLGDTSVLSSKRPMYGDFTGLPSDPIEALNLVHQSEYAFAQLSADDKAKYNNDWRR 121
+H +G + ++ YG+ +G + +VH E +FAQ+ ++ +R+
Sbjct 113 IHTFHIGKNASITYTEKHYGEGSGTGGKILNPTTVVHMEEGSFAQMDMSQIKGVDSTFRK 172
Query 122 WFADLLSG-RDNLSEKL---SSVVPNSDVEK--EGADSVVE 156
A+L +G + ++EKL + +SDV G DSVV+
Sbjct 173 TEANLAAGAKLVINEKLMTHGTQTAHSDVTVNLNGEDSVVQ 213
Lambda K H a alpha
0.311 0.130 0.371 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 434602183812