bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_3
Length=240
Score E
Sequences producing significant alignments: (Bits) Value
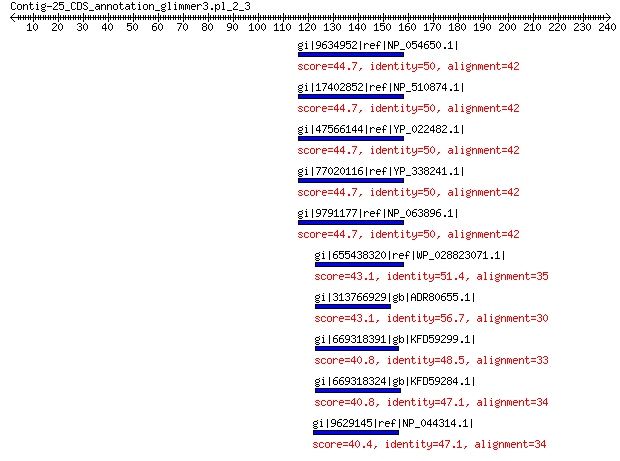
gi|9634952|ref|NP_054650.1| structural protein 44.7 0.016
gi|17402852|ref|NP_510874.1| capsid protein VP2-related protein 44.7 0.016
gi|47566144|ref|YP_022482.1| structural protein 44.7 0.017
gi|77020116|ref|YP_338241.1| putative minor spike 44.7 0.017
gi|9791177|ref|NP_063896.1| capsid protein VP2-related protein 44.7 0.017
gi|655438320|ref|WP_028823071.1| hypothetical protein 43.1 0.049
gi|313766929|gb|ADR80655.1| putative minor capsid protein 43.1 0.070
gi|669318391|gb|KFD59299.1| hypothetical protein M514_28523 40.8 0.11
gi|669318324|gb|KFD59284.1| hypothetical protein M514_28537 40.8 0.47
gi|9629145|ref|NP_044314.1| VP2 40.4 0.64
>gi|9634952|ref|NP_054650.1| structural protein [Chlamydia phage 2]
gi|7406592|emb|CAB85592.1| structural protein [Chlamydia phage 2]
Length=186
Score = 44.7 bits (104), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 21/43 (49%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 116 ASAYGMSKE-TAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
A+A +++E A+ E M NTAY+R++ DM+KAGLNP + F G
Sbjct 42 ATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLNPMLAFSKG 84
>gi|17402852|ref|NP_510874.1| capsid protein VP2-related protein [Guinea pig Chlamydia phage]
Length=186
Score = 44.7 bits (104), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 21/43 (49%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 116 ASAYGMSKE-TAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
A+A +++E A+ E M NTAY+R++ DM+KAGLNP + F G
Sbjct 42 ATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLNPMLAFSKG 84
>gi|47566144|ref|YP_022482.1| structural protein [Chlamydia phage 3]
gi|47522479|emb|CAD79480.1| structural protein [Chlamydia phage 3]
Length=186
Score = 44.7 bits (104), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 21/43 (49%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 116 ASAYGMSKE-TAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
A+A +++E A+ E M NTAY+R++ DM+KAGLNP + F G
Sbjct 42 ATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLNPMLAFSKG 84
>gi|77020116|ref|YP_338241.1| putative minor spike [Chlamydia phage 4]
gi|59940015|gb|AAX12544.1| putative minor spike [Chlamydia phage 4]
Length=186
Score = 44.7 bits (104), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 21/43 (49%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 116 ASAYGMSKE-TAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
A+A +++E A+ E M NTAY+R++ DM+KAGLNP + F G
Sbjct 42 ATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLNPMLAFSKG 84
>gi|9791177|ref|NP_063896.1| capsid protein VP2-related protein [Chlamydia pneumoniae phage
CPAR39]
gi|7190962|gb|AAF39722.1| capsid protein VP2-related protein [Chlamydia pneumoniae phage
CPAR39]
Length=186
Score = 44.7 bits (104), Expect = 0.017, Method: Compositional matrix adjust.
Identities = 21/43 (49%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 116 ASAYGMSKE-TAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
A+A +++E A+ E M NTAY+R++ DM+KAGLNP + F G
Sbjct 42 ATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLNPMLAFSKG 84
>gi|655438320|ref|WP_028823071.1| hypothetical protein [Proteobacteria bacterium JGI 0000113-P07]
Length=186
Score = 43.1 bits (100), Expect = 0.049, Method: Compositional matrix adjust.
Identities = 18/35 (51%), Positives = 27/35 (77%), Gaps = 0/35 (0%)
Query 123 KETAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
++ A+ E M NTAY+RS++DMQ+AGLNP + +G
Sbjct 30 RQMAFQERMSNTAYQRSMADMQQAGLNPILAAKSG 64
>gi|313766929|gb|ADR80655.1| putative minor capsid protein [Uncultured Microviridae]
Length=220
Score = 43.1 bits (100), Expect = 0.070, Method: Compositional matrix adjust.
Identities = 17/30 (57%), Positives = 25/30 (83%), Gaps = 0/30 (0%)
Query 123 KETAYSEAMENTAYRRSVSDMQKAGLNPAV 152
++ A+ E M NTAY+RS++DM+KAGLNP +
Sbjct 29 RQMAFQERMSNTAYQRSMADMRKAGLNPIL 58
>gi|669318391|gb|KFD59299.1| hypothetical protein M514_28523, partial [Trichuris suis]
Length=95
Score = 40.8 bits (94), Expect = 0.11, Method: Compositional matrix adjust.
Identities = 16/33 (48%), Positives = 24/33 (73%), Gaps = 0/33 (0%)
Query 123 KETAYSEAMENTAYRRSVSDMQKAGLNPAVIFG 155
K+ A+ E M NTAY+R+ +D++ AGLNP + G
Sbjct 7 KQQAFQERMSNTAYQRAAADLKAAGLNPLLALG 39
>gi|669318324|gb|KFD59284.1| hypothetical protein M514_28537 [Trichuris suis]
Length=216
Score = 40.8 bits (94), Expect = 0.47, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 25/34 (74%), Gaps = 0/34 (0%)
Query 123 KETAYSEAMENTAYRRSVSDMQKAGLNPAVIFGA 156
K+ A+ E M NTAY+R+ +D++ AGLNP + G+
Sbjct 83 KQQAFQERMSNTAYQRAAADLKAAGLNPLLALGS 116
>gi|9629145|ref|NP_044314.1| VP2 [Chlamydia phage 1]
gi|139201|sp|P19193.2|H_BPCHP RecName: Full=Minor spike protein H; AltName: Full=H protein;
AltName: Full=Pilot protein; AltName: Full=Protein VP2; Short=VP2
[Chlamydia phage 1]
gi|93810|pir||JU0346 capsid protein VP2 - Chlamydophila psittaci phage Chp1
gi|217764|dbj|BAA00505.1| VP2 [Chlamydia phage 1]
Length=263
Score = 40.4 bits (93), Expect = 0.64, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 24/34 (71%), Gaps = 0/34 (0%)
Query 122 SKETAYSEAMENTAYRRSVSDMQKAGLNPAVIFG 155
+++ A+ E M +TA RR V D++KAGLNP + G
Sbjct 86 NRQMAFQERMSSTAVRRHVEDLKKAGLNPLLALG 119
Lambda K H a alpha
0.312 0.127 0.371 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1023805680780