bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_8
Length=286
Score E
Sequences producing significant alignments: (Bits) Value
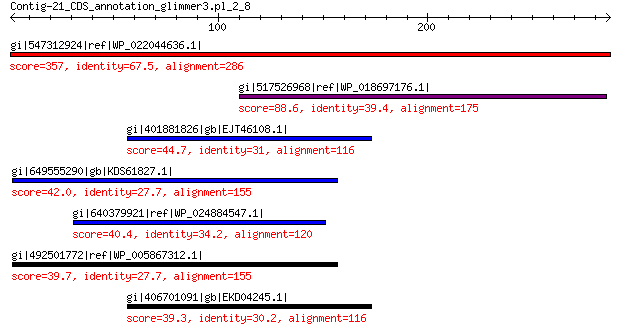
gi|547312924|ref|WP_022044636.1| putative uncharacterized protein 357 4e-119
gi|517526968|ref|WP_018697176.1| hypothetical protein 88.6 5e-17
gi|401881826|gb|EJT46108.1| hypothetical protein A1Q1_05319 44.7 0.061
gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 42.0 0.29
gi|640379921|ref|WP_024884547.1| hypothetical protein 40.4 1.4
gi|492501772|ref|WP_005867312.1| hypothetical protein 39.7 1.6
gi|406701091|gb|EKD04245.1| hypothetical protein A1Q2_01464 39.3 3.1
>gi|547312924|ref|WP_022044636.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208405|emb|CCZ76640.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=328
Score = 357 bits (915), Expect = 4e-119, Method: Compositional matrix adjust.
Identities = 193/289 (67%), Positives = 232/289 (80%), Gaps = 15/289 (5%)
Query 1 MNKPSRQVAKWRSAGIAPQAVFGNSPGGAGIVTDASTPNSQTPMGSSDFNFVTTIAERQR 60
MNKPSRQVAKWRSAGIAPQAVFGNSPGGAGI TDAS+PNSQTPMGSSDFNFVTTIAERQR
Sbjct 49 MNKPSRQVAKWRSAGIAPQAVFGNSPGGAGIATDASSPNSQTPMGSSDFNFVTTIAERQR 108
Query 61 MKNEKAIADATVNKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAVDEIN 120
MKNEKAIADATV+KLNAEA KLRGDTKDP VTKD Q+LEFDWN+VKKQRE+VQL VDEI+
Sbjct 109 MKNEKAIADATVDKLNAEAGKLRGDTKDPKVTKDLQQLEFDWNIVKKQRERVQLDVDEID 168
Query 121 KEFQRAINEADVQIKHGLYSETLAKIDKLIADKDVSEEMKQNLQKQRDLIESQISATQAQ 180
KEF+RA+NEAD+QIK G+YSETL+KIDKLIADK+VSEEMKQNLQKQRDLI++QI +T+AQ
Sbjct 169 KEFRRAVNEADLQIKRGIYSETLSKIDKLIADKEVSEEMKQNLQKQRDLIDAQIDSTKAQ 228
Query 181 TDLIKaqtsatqaqtETENALRDGRIKLTEREANKILADIGLSEARSLNE-YESLIKAMT 239
T L K AQT+TE+ LRDGR+KLT + +++L+ GL++ R E YE+ ++ +
Sbjct 229 TGLSK-------AQTKTEDTLRDGRVKLTGAQTSELLSMAGLNDVRRDREKYETFLR-LL 280
Query 240 GTQPASSLWGYIDRLIARGDSRLGGYENA--SDLREKLARALVRYIKAD 286
AS+ + R+I +L G+ NA SD + K+ + I +D
Sbjct 281 DIDDASNGAEFAQRII----RQLFGHMNADLSDYKRKMISDYLEKIWSD 325
>gi|517526968|ref|WP_018697176.1| hypothetical protein [Alistipes onderdonkii]
Length=364
Score = 88.6 bits (218), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 69/191 (36%), Positives = 101/191 (53%), Gaps = 26/191 (14%)
Query 110 EQVQLAVDEINKEFQRAINEADV------------QIKHGLYSETLAKIDKLIAD--KDV 155
+Q + V E+ +F A +D+ Q K+ L S + D+L D KD
Sbjct 184 KQFKATVAEVESQFAEAQALSDLAERNARIESIWAQAKNSLASAAKSDADRLYLDFMKDA 243
Query 156 SEEMKQNLQKQRDLIESQISATQAQTDLIKaqtsatqaqtETENALRDGRIKLTEREANK 215
+ R+ ++SQ + QAQ + + AQ ETE+ALR GRIKLTE +A
Sbjct 244 N----------RENVQSQTALNQAQAGTATSSAALMDAQRETEDALRSGRIKLTEEQARA 293
Query 216 ILADIGLSEARSLNEYESLIKAMTGTQPASSLWGYIDRLIARGDSRL-GGYENASD-LRE 273
LA GLSEAR+ EY LI+A+T T+ A+SLWG +DR + + ++ L GG + +D LR
Sbjct 294 ALASAGLSEARAGREYNELIEALTNTRSANSLWGIVDRYVRKTEAILPGGPQGKADELRF 353
Query 274 KLARALVRYIK 284
L +A+ Y K
Sbjct 354 ALIKAISNYSK 364
>gi|401881826|gb|EJT46108.1| hypothetical protein A1Q1_05319 [Trichosporon asahii var. asahii
CBS 2479]
Length=936
Score = 44.7 bits (104), Expect = 0.061, Method: Compositional matrix adjust.
Identities = 36/117 (31%), Positives = 62/117 (53%), Gaps = 8/117 (7%)
Query 57 ERQRMKNEKAIADATVNKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAV 116
ER+R+K E A DA V +LNAE E+LR ++P+V + D + V + +EQ+ A+
Sbjct 632 ERRRLKKEVADRDAQVERLNAETERLREQPQEPSVRSEPG----DSSAVAELQEQLDAAM 687
Query 117 DEINKEFQRAINEADVQIKHGLYSETLAKIDKLIAD-KDVSEEMKQNLQKQRDLIES 172
+++ I D++ +H + L + +AD D E+ ++L RD I+S
Sbjct 688 SKLSS---LEIQHQDLEAEHDDATAELEHTRQKLADTNDALEKASEDLYAARDAIDS 741
>gi|649555290|gb|KDS61827.1| hypothetical protein M095_3809 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649557306|gb|KDS63785.1| hypothetical protein M095_3404 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649559158|gb|KDS65545.1| hypothetical protein M096_4689 [Parabacteroides distasonis str.
3999B T(B) 6]
gi|649560567|gb|KDS66875.1| hypothetical protein M095_2448 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561016|gb|KDS67303.1| hypothetical protein M095_2410 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649562727|gb|KDS68911.1| hypothetical protein M096_3341 [Parabacteroides distasonis str.
3999B T(B) 6]
Length=288
Score = 42.0 bits (97), Expect = 0.29, Method: Compositional matrix adjust.
Identities = 43/187 (23%), Positives = 81/187 (43%), Gaps = 37/187 (20%)
Query 2 NKPSRQVAKWRSAGIAPQAVFGNSPGGAGIVTDASTP-------NSQTPMGSSDFNFVTT 54
N P++Q+A+ R+AG+ P V+GN G + STP N+ T +N +
Sbjct 68 NSPTQQMARIRAAGLNPNLVYGNGVTGN---SAGSTPQYEPAKFNAPTMQAYRGWNLGIS 124
Query 55 IAERQRMKNEKAIADATVNKLNAEAEKLRGD-----TKDPNVTKDSQRLEFDWNLVKKQR 109
A Q + A V+ + A+ +R T+ N+ + R EFD N+ K+ +
Sbjct 125 DATSQYLAYR--TVKAQVDNMEAQNSLIRQQTATEATRQANIAASTSRSEFDLNMAKELK 182
Query 110 E-QVQLAVDEINK-------------------EFQRAINEADVQIKHGLYSETLAKIDKL 149
+ V A+ E+N+ E +A+ + +++ + Y + L + +L
Sbjct 183 DVSVSSAIAEMNQKQAVAAQGWTKANREVVQYELDKALFDNKIKLNNEKYLKALQSVRQL 242
Query 150 IADKDVS 156
D D++
Sbjct 243 TQDNDIN 249
>gi|640379921|ref|WP_024884547.1| hypothetical protein [Streptomyces sp. CNH189]
Length=1293
Score = 40.4 bits (93), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 41/138 (30%), Positives = 70/138 (51%), Gaps = 19/138 (14%)
Query 31 IVTDASTPN---SQTPMGSSDFNFVTTIAERQRM------KNEKAIADAT--VNKLNAEA 79
++T+A + ++ + +D T+AE +R+ K EK IADAT +L AEA
Sbjct 919 LITEARSEAERLTEDTITETDRLRTETVAEAERVRSESVAKAEKLIADATGDAERLRAEA 978
Query 80 EKLRGDTKD--PNVTKDSQRLEFDWN-----LVKKQREQVQLAVDEINKEFQRAINEADV 132
+ G + + +D++R++ D LV RE+ + +DE KE + +EA
Sbjct 979 AETVGSAQQHAERIRRDAERVKTDAETEAERLVSGAREEAERTLDEARKEANKRRSEAAE 1038
Query 133 QIKHGLYSETLAKIDKLI 150
Q+ L +ET A+ DKL+
Sbjct 1039 QV-DTLITETTAEADKLL 1055
>gi|492501772|ref|WP_005867312.1| hypothetical protein [Parabacteroides distasonis]
gi|409230405|gb|EKN23269.1| hypothetical protein HMPREF1059_03254 [Parabacteroides distasonis
CL09T03C24]
Length=288
Score = 39.7 bits (91), Expect = 1.6, Method: Compositional matrix adjust.
Identities = 43/187 (23%), Positives = 80/187 (43%), Gaps = 37/187 (20%)
Query 2 NKPSRQVAKWRSAGIAPQAVFGNSPGGAGIVTDASTP-------NSQTPMGSSDFNFVTT 54
N P++Q+A+ R+AG+ P V+GN G + STP N+ T +N +
Sbjct 68 NSPTQQMARIRAAGLNPNLVYGNGVTGN---SSGSTPQYEPAKFNAPTMQAYRGWNLGIS 124
Query 55 IAERQRMKNEKAIADATVNKLNAEAEKLRGD-----TKDPNVTKDSQRLEFDWNLVKKQR 109
A Q + A V+ + A+ +R TK N+ + R EFD N+ K+ +
Sbjct 125 DAISQFLAYR--TVKAQVDNMEAQNSLIRQQTATEATKQANIAASTSRSEFDLNMAKELK 182
Query 110 E-QVQLAVDEINK-------------------EFQRAINEADVQIKHGLYSETLAKIDKL 149
+ V A+ ++N+ E +A+ + +++ + Y L + +L
Sbjct 183 DVSVSSAIADMNQKQAGAAQGWTKANREVIQYELDKALFDNKIKLSNQEYLRVLQSVRQL 242
Query 150 IADKDVS 156
D D++
Sbjct 243 QQDNDIN 249
>gi|406701091|gb|EKD04245.1| hypothetical protein A1Q2_01464 [Trichosporon asahii var. asahii
CBS 8904]
Length=936
Score = 39.3 bits (90), Expect = 3.1, Method: Compositional matrix adjust.
Identities = 35/118 (30%), Positives = 62/118 (53%), Gaps = 10/118 (8%)
Query 57 ERQRMKNEKAIADATVNKLNAEAEKLRGDTKDPNVTKDSQRLEFDWNLVKKQREQVQLAV 116
ER+R+K E A DA V +LNAE E+L ++P+V + D + V + +EQ+ A
Sbjct 632 ERRRLKKEVADRDAQVERLNAETERLHEQPQEPSVRSEPG----DSSAVAELQEQLDAAT 687
Query 117 DEINK-EFQRAINEADVQIKHGLYSETLAKIDKLIADKDVS-EEMKQNLQKQRDLIES 172
+++ E Q D++ +H + L + +AD + + E+ ++L RD I+S
Sbjct 688 SKLSSLELQ----HQDLEAEHDDATAELEHTRQKLADTNNALEKASEDLYAARDAIDS 741
Lambda K H a alpha
0.311 0.127 0.342 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1457072974140