bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-21_CDS_annotation_glimmer3.pl_2_4
Length=60
Score E
Sequences producing significant alignments: (Bits) Value
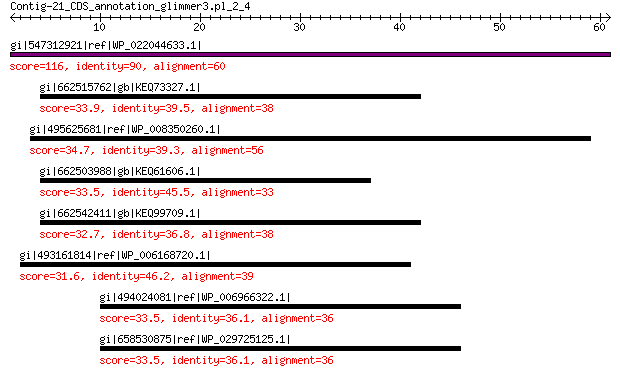
gi|547312921|ref|WP_022044633.1| hypothetical protein 116 3e-32
gi|662515762|gb|KEQ73327.1| hypothetical protein M436DRAFT_63642 33.9 3.3
gi|495625681|ref|WP_008350260.1| NHL repeat protein 34.7 4.3
gi|662503988|gb|KEQ61606.1| hypothetical protein M437DRAFT_67069 33.5 4.6
gi|662542411|gb|KEQ99709.1| hypothetical protein AUEXF2481DRAFT_... 32.7 6.8
gi|493161814|ref|WP_006168720.1| transposase 31.6 7.2
gi|494024081|ref|WP_006966322.1| integral membrane sensor signal... 33.5 7.9
gi|658530875|ref|WP_029725125.1| hypothetical protein 33.5 9.5
>gi|547312921|ref|WP_022044633.1| hypothetical protein [Alistipes finegoldii CAG:68]
gi|524208441|emb|CCZ76637.1| unknown [Alistipes finegoldii CAG:68]
Length=60
Score = 116 bits (290), Expect = 3e-32, Method: Compositional matrix adjust.
Identities = 54/60 (90%), Positives = 59/60 (98%), Gaps = 0/60 (0%)
Query 1 MEIQKEYIIVVNGRPYFSVVDVEHLSAVIDDAKARFGADSKIEVVMQTTEPYAPGKDNGN 60
MEIQKEYI+VVNGRPYFSVVDV+HLSAVIDDAKARFGADSKI+V +QTTEPYAPGK+NGN
Sbjct 1 MEIQKEYILVVNGRPYFSVVDVKHLSAVIDDAKARFGADSKIDVFLQTTEPYAPGKNNGN 60
>gi|662515762|gb|KEQ73327.1| hypothetical protein M436DRAFT_63642 [Aureobasidium pullulans
var. namibiae CBS 147.97]
Length=174
Score = 33.9 bits (76), Expect = 3.3, Method: Compositional matrix adjust.
Identities = 15/38 (39%), Positives = 23/38 (61%), Gaps = 4/38 (11%)
Query 4 QKEYIIVVNGRPYFSVVDVEHLSAVIDDAKARFGADSK 41
+ EY+I+ N RPY HL+A+I+D + RF D +
Sbjct 111 KSEYLILANQRPYRRT----HLAAMIEDVETRFTIDEQ 144
>gi|495625681|ref|WP_008350260.1| NHL repeat protein, partial [Thiovulum sp. ES]
gi|394446527|gb|EJF07348.1| NHL repeat protein, partial [Thiovulum sp. ES]
Length=4349
Score = 34.7 bits (78), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 22/59 (37%), Positives = 30/59 (51%), Gaps = 3/59 (5%)
Query 3 IQKEYIIVVNGRPYFSVVDV---EHLSAVIDDAKARFGADSKIEVVMQTTEPYAPGKDN 58
+ K + I+++G P + VDV E V DD D KIEV+ EPYA G+ N
Sbjct 964 VPKTHEILISGSPNYGNVDVGEHEFTITVSDDTGRSIEYDDKIEVIDVNFEPYASGESN 1022
>gi|662503988|gb|KEQ61606.1| hypothetical protein M437DRAFT_67069 [Aureobasidium melanogenum
CBS 110374]
Length=157
Score = 33.5 bits (75), Expect = 4.6, Method: Compositional matrix adjust.
Identities = 15/33 (45%), Positives = 21/33 (64%), Gaps = 4/33 (12%)
Query 4 QKEYIIVVNGRPYFSVVDVEHLSAVIDDAKARF 36
+ EY+I+ N RPY HL+A+I+DA RF
Sbjct 94 KSEYLILANQRPYRRA----HLAAMIEDADTRF 122
>gi|662542411|gb|KEQ99709.1| hypothetical protein AUEXF2481DRAFT_1198 [Aureobasidium subglaciale
EXF-2481]
Length=157
Score = 32.7 bits (73), Expect = 6.8, Method: Compositional matrix adjust.
Identities = 14/38 (37%), Positives = 23/38 (61%), Gaps = 4/38 (11%)
Query 4 QKEYIIVVNGRPYFSVVDVEHLSAVIDDAKARFGADSK 41
+ EY+++ N RPY HL+A+I+DA RF + +
Sbjct 94 KSEYLVLANQRPYRR----SHLAAMIEDADTRFTTEEQ 127
>gi|493161814|ref|WP_006168720.1| transposase [Natrialba chahannaoensis]
gi|445643072|gb|ELY96127.1| transposase [Natrialba chahannaoensis JCM 10990]
Length=83
Score = 31.6 bits (70), Expect = 7.2, Method: Compositional matrix adjust.
Identities = 18/39 (46%), Positives = 22/39 (56%), Gaps = 0/39 (0%)
Query 2 EIQKEYIIVVNGRPYFSVVDVEHLSAVIDDAKARFGADS 40
E + E IIV++G PYF V L+A D A RF A S
Sbjct 34 EFEDELIIVLDGAPYFQASAVTDLAARDDLALVRFPAYS 72
>gi|494024081|ref|WP_006966322.1| integral membrane sensor signal transduction histidine kinase
ZraS [Desulfotignum phosphitoxidans]
gi|474488013|gb|EMS79232.1| integral membrane sensor signal transduction histidine kinase
ZraS [Desulfotignum phosphitoxidans DSM 13687]
Length=587
Score = 33.5 bits (75), Expect = 7.9, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 24/36 (67%), Gaps = 0/36 (0%)
Query 10 VVNGRPYFSVVDVEHLSAVIDDAKARFGADSKIEVV 45
VVN PY+S++D EHL+ ++ + K FG + + ++
Sbjct 101 VVNELPYYSLIDNEHLTKILINLKLGFGGFTDLSII 136
>gi|658530875|ref|WP_029725125.1| hypothetical protein, partial [Desulfotignum balticum]
Length=557
Score = 33.5 bits (75), Expect = 9.5, Method: Composition-based stats.
Identities = 13/36 (36%), Positives = 24/36 (67%), Gaps = 0/36 (0%)
Query 10 VVNGRPYFSVVDVEHLSAVIDDAKARFGADSKIEVV 45
VVN PY+S++D EHL+ ++ + K FG + + ++
Sbjct 71 VVNELPYYSLIDNEHLTKILINLKLGFGGFTDLSII 106
Lambda K H a alpha
0.315 0.134 0.375 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 430540141590