bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_6
Length=367
Score E
Sequences producing significant alignments: (Bits) Value
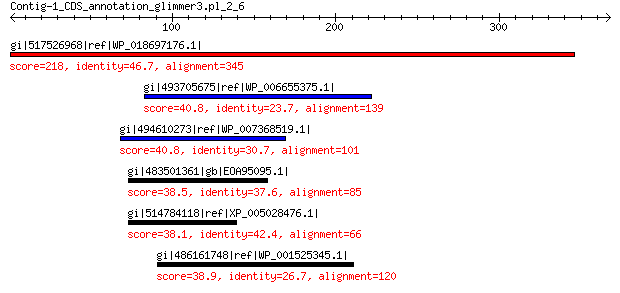
gi|517526968|ref|WP_018697176.1| hypothetical protein 218 1e-63
gi|493705675|ref|WP_006655375.1| sugar nucleotidyltransferase 40.8 0.88
gi|494610273|ref|WP_007368519.1| hypothetical protein 40.8 1.4
gi|483501361|gb|EOA95095.1| Tumor susceptibility gene 101 protein 38.5 6.4
gi|514784118|ref|XP_005028476.1| PREDICTED: tumor susceptibility... 38.1 7.9
gi|486161748|ref|WP_001525345.1| conjugal transfer protein TraI 38.9 8.2
>gi|517526968|ref|WP_018697176.1| hypothetical protein [Alistipes onderdonkii]
Length=364
Score = 218 bits (555), Expect = 1e-63, Method: Compositional matrix adjust.
Identities = 161/350 (46%), Positives = 219/350 (63%), Gaps = 12/350 (3%)
Query 1 MSALVTsaiiagasglaaaggssiaasKMNSRAEKYNRWALKEQQRYQKEYADYMAQLEA 60
MSALVTSAII GA+ L +AG S+I+A KMN RA +Y+ K Q+ +QKEY+ Y+++LEA
Sbjct 1 MSALVTSAIIGGAAALGSAGVSAISAGKMNRRAVRYSVAENKRQRAFQKEYSKYLSELEA 60
Query 61 QQNNLYWDKYNSPAAQRRARVAAGFTPYADVGGIQTSSVDPGSYGGSTPSAQSFTQPGGI 120
+QN YW+KYNSP+AQR AR+ AG +P+AD G+Q VDPGSY GS+PS+Q FTQPGG
Sbjct 61 KQNQEYWNKYNSPSAQRIARMKAGMSPFADESGVQAMGVDPGSYSGSSPSSQPFTQPGGN 120
Query 121 PSSPLVGAFGNATQQTLSALQAEANIELTKSQALKTRAETTGLENTNSMFDVVKSIADEE 180
SPL AF + QQ LSA QAEANI+LT + KT+AE + NS+F K+ A+ +
Sbjct 121 AMSPLSPAFASGVQQVLSARQAEANIQLTDANTAKTQAEAVKAQQENSLFAFTKAAAESD 180
Query 181 LTSKRFSNILKEVEVKYAEVNAITDLDAKQAKIAEINASALERLASAAKTDADRITVELL 240
SK+F + EVE ++AE A++DL + A+I I A A LASAAK+DADR+ ++ +
Sbjct 181 ALSKQFKATVAEVESQFAEAQALSDLAERNARIESIWAQAKNSLASAAKSDADRLYLDFM 240
Query 241 RDAQKRsleagaslaeaqaaTEPHRALNLKQDTLLKMAQEETEQLLRSQKFELTRQQARA 300
+DA + ++++ +L +AQA T A L AQ ETE LRS + +LT +QARA
Sbjct 241 KDANRENVQSQTALNQAQAGTATSSA-------ALMDAQRETEDALRSGRIKLTEEQARA 293
Query 301 AAISFVQERILTYRQAEELARYLANIHDPKNMWDGIWRIVS-----LPAG 345
A S R+ EL L N ++W + R V LP G
Sbjct 294 ALASAGLSEARAGREYNELIEALTNTRSANSLWGIVDRYVRKTEAILPGG 343
>gi|493705675|ref|WP_006655375.1| sugar nucleotidyltransferase [Helicobacter canadensis]
gi|253510733|gb|EES89392.1| conserved hypothetical protein [Helicobacter canadensis MIT 98-5491]
gi|313130566|gb|EFR48183.1| hypothetical protein HCMG_00356 [Helicobacter canadensis MIT
98-5491]
Length=252
Score = 40.8 bits (94), Expect = 0.88, Method: Compositional matrix adjust.
Identities = 33/139 (24%), Positives = 60/139 (43%), Gaps = 2/139 (1%)
Query 83 AGFTPYADVGGIQTSSVDPGSYGGSTPSAQSFTQPGGIPSSPLVGAFGNATQQTLSALQA 142
AG A VGG + Y T + + F + S+ +V A Q S +Q
Sbjct 44 AGIIEIAVVGGYLQEVLR--DYLSKTYALKEFFKNPNFDSTNMVATLFCARQWIESCIQH 101
Query 143 EANIELTKSQALKTRAETTGLENTNSMFDVVKSIADEELTSKRFSNILKEVEVKYAEVNA 202
+ ++ ++ + + ++ L+NT+ F ++ EL KRF N L++VE + N
Sbjct 102 KEDLLISYADIIYSKEIVEKLKNTDVPFGIIVDKNWRELWEKRFDNPLEDVETLKIKENR 161
Query 203 ITDLDAKQAKIAEINASAL 221
+ +L K +EI +
Sbjct 162 VVELGKKPKDYSEIEGQYI 180
>gi|494610273|ref|WP_007368519.1| hypothetical protein [Prevotella multiformis]
gi|324988545|gb|EGC20508.1| hypothetical protein HMPREF9141_0987 [Prevotella multiformis
DSM 16608]
Length=437
Score = 40.8 bits (94), Expect = 1.4, Method: Compositional matrix adjust.
Identities = 31/101 (31%), Positives = 48/101 (48%), Gaps = 3/101 (3%)
Query 68 DKYNSPAAQRRARVAAGFTPYADVGGIQTSSVDPGSYGGSTPSAQSFTQPGGIPSSPLVG 127
++YNSPAAQ + AG PY G +QT + P Q Q +P++ +
Sbjct 130 NQYNSPAAQMQRYTDAGINPYIAAGNVQTGNAQSALQSAPAPQ-QHVAQV--MPATGMGD 186
Query 128 AFGNATQQTLSALQAEANIELTKSQALKTRAETTGLENTNS 168
A N+ Q + + A +L +QA KT AE + ++ NS
Sbjct 187 AVQNSFAQIGNVISQFAQNQLALAQAKKTDAEASWIDRLNS 227
>gi|483501361|gb|EOA95095.1| Tumor susceptibility gene 101 protein, partial [Anas platyrhynchos]
Length=377
Score = 38.5 bits (88), Expect = 6.4, Method: Compositional matrix adjust.
Identities = 32/105 (30%), Positives = 42/105 (40%), Gaps = 21/105 (20%)
Query 73 PAAQRRARVAAGFTPYADVGGIQTSSVDPGSYGGST---------PSAQSFTQPGGI--- 120
P R V+AG+ PY G TS V PG GG T PS ++ PGG+
Sbjct 125 PPVFSRPTVSAGYPPYQATGPPTTSYV-PGMPGGITPYPPGSTANPSYPNYPYPGGVPFP 183
Query 121 --------PSSPLVGAFGNATQQTLSALQAEANIELTKSQALKTR 157
PS P V G + T+S A++ S L+ R
Sbjct 184 ATTSVQYYPSQPPVTTVGPSRDGTISEDTIRASLISAVSDKLRWR 228
>gi|514784118|ref|XP_005028476.1| PREDICTED: tumor susceptibility gene 101 protein [Anas platyrhynchos]
Length=351
Score = 38.1 bits (87), Expect = 7.9, Method: Compositional matrix adjust.
Identities = 28/87 (32%), Positives = 35/87 (40%), Gaps = 22/87 (25%)
Query 73 PAAQRRARVAAGFTPYADVGGIQTSSVDPGSYGGSTP----------SAQSFTQPGGI-- 120
P R V+AG+ PY G TS V PG GG TP S ++ PGG+
Sbjct 98 PPVFSRPTVSAGYPPYQATGPPTTSYV-PGMPGGITPYPPGSTANPSSYPNYPYPGGVPF 156
Query 121 ---------PSSPLVGAFGNATQQTLS 138
PS P V G + T+S
Sbjct 157 PATTSVQYYPSQPPVTTVGPSRDGTIS 183
>gi|486161748|ref|WP_001525345.1| conjugal transfer protein TraI [Salmonella enterica]
gi|205321805|gb|EDZ09644.1| conjugative transfer relaxase protein TraI [Salmonella enterica
subsp. enterica serovar Saintpaul str. SARA29]
Length=1765
Score = 38.9 bits (89), Expect = 8.2, Method: Compositional matrix adjust.
Identities = 32/122 (26%), Positives = 61/122 (50%), Gaps = 6/122 (5%)
Query 91 VGGIQTSSVDPGSYGGSTPSAQSFTQPGGIPSSPLVGA-FGNATQQTLSALQAEANIELT 149
+G QT + PG G T +S QPGG+ S+ + + A T+SAL +E ++ T
Sbjct 289 IGLAQTRAEPPGPVVGGTEGIRSTGQPGGVSSAQISESDVQKAVSDTISAL-SEKKVQFT 347
Query 150 KSQALKTRAETTG-LENTNSMFDVVKSIADEELTSKRFSNILKEVEVKYAEVNAITDLDA 208
S+ L A T G L + + +F+ ++ D + +R + +E + ++++ + +L
Sbjct 348 WSEML---AGTVGRLPSVSGLFEQARAGIDAAIEEQRLIPLDREKGIFTSDIHLLNELSV 404
Query 209 KQ 210
Q
Sbjct 405 HQ 406
Lambda K H a alpha
0.312 0.125 0.339 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2236817440716