bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
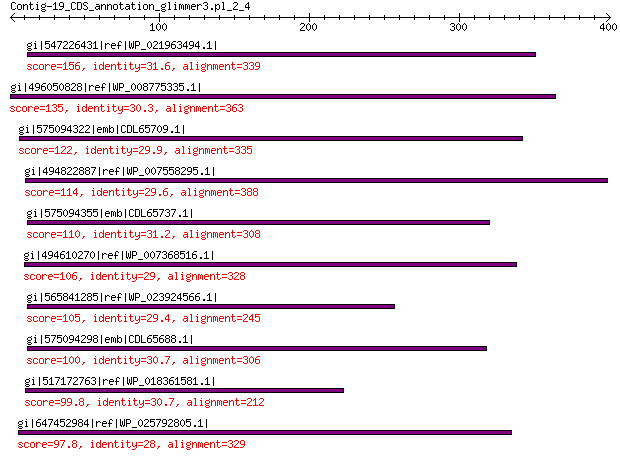
Query= Contig-19_CDS_annotation_glimmer3.pl_2_4
Length=400
Score E
Sequences producing significant alignments: (Bits) Value
gi|547226431|ref|WP_021963494.1| predicted protein 156 4e-39
gi|496050828|ref|WP_008775335.1| hypothetical protein 135 7e-32
gi|575094322|emb|CDL65709.1| unnamed protein product 122 3e-27
gi|494822887|ref|WP_007558295.1| hypothetical protein 114 1e-24
gi|575094355|emb|CDL65737.1| unnamed protein product 110 2e-23
gi|494610270|ref|WP_007368516.1| hypothetical protein 106 5e-22
gi|565841285|ref|WP_023924566.1| hypothetical protein 105 1e-21
gi|575094298|emb|CDL65688.1| unnamed protein product 100 5e-20
gi|517172763|ref|WP_018361581.1| hypothetical protein 99.8 2e-19
gi|647452984|ref|WP_025792805.1| hypothetical protein 97.8 6e-19
>gi|547226431|ref|WP_021963494.1| predicted protein [Prevotella sp. CAG:1185]
gi|524103383|emb|CCY83995.1| predicted protein [Prevotella sp. CAG:1185]
Length=498
Score = 156 bits (395), Expect = 4e-39, Method: Compositional matrix adjust.
Identities = 107/350 (31%), Positives = 170/350 (49%), Gaps = 52/350 (15%)
Query 12 CLHPRVIKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYL 71
C HPR ++NKYTG+ + V CG C+ C+ +A K C ++ + KYC F TLTYS +Y+
Sbjct 11 CYHPRHVQNKYTGEVIQVGCGVCKACLKRRADKMSFLCAIEEQSHKYCMFATLTYSNDYV 70
Query 72 PVGEFYKGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYF----DFDTQLSWESAQL 127
P Y E+R L R + Y + + + ++ + +Y+ DT + +A+
Sbjct 71 P--RMYPEVDNELR---LVRWYSYCDRLNEK-GKLMTVDYDYWHKCPSLDTYVLMLTAKC 124
Query 128 LQKKTHLHYTSFPDGRRIYNRPYMENLIGYLNY---RDMQLFFKRLNQNIRSVTNEKIYY 184
NL GYL+Y RD QLF KR+ +N+ ++EKI Y
Sbjct 125 -------------------------NLDGYLSYTSKRDAQLFLKRVRKNLSKYSDEKIRY 159
Query 185 YVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSWRFGDTDTQPVWSSASCYVAGYV 244
Y+V EYGP TFR H+H+L F+D + ++ + + + ++W+FG D + YVA YV
Sbjct 160 YIVSEYGPKTFRAHYHVLFFYDEVKTQKVMSKVIRQAWQFGRVDCSLSRGKCNSYVARYV 219
Query 245 NSTACLPDFYKNFSHIKPFG----RFSMHFAESAFNEVFKPQEDEEIFSLFYDGRMLELN 300
N CLP F + S KPF RF++ +S E++K D+ I+ + E+N
Sbjct 220 NCNYCLPRFLGDMS-TKPFSCHSIRFALGIHQSQKEEIYKGSVDDFIY------QSGEIN 272
Query 301 GKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATALSNIPHVLAKFGF 350
G P R+ +P K K S D + + + + V + G+
Sbjct 273 GNYVEFMPWRNLSCTFFP---KCKGYSRKSDSELWQSYNILREVRSAIGY 319
>gi|496050828|ref|WP_008775335.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448892|gb|EEO54683.1| hypothetical protein BSCG_01608 [Bacteroides sp. 2_2_4]
Length=497
Score = 135 bits (340), Expect = 7e-32, Method: Compositional matrix adjust.
Identities = 110/367 (30%), Positives = 163/367 (44%), Gaps = 53/367 (14%)
Query 1 MINNRSKAYGSCLHPRVIKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCE 60
M+ N + CLHP+ I N YT + + VPCG C+ C K + +C+++ +K+
Sbjct 1 MVQN---PFCKCLHPKRIMNPYTKESMVVPCGHCQACTLAKNSRYAFQCDLESYTAKHTL 57
Query 61 FITLTYSTEYLPVGEFYKGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQL 120
FITLTY+ ++P F S E + C + D +T
Sbjct 58 FITLTYANRFIPRAMFVD--SIERPYGC-----------------------DLIDKETGE 92
Query 121 SWESAQLLQKKTHLHYTSFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNI-RSVTN 179
A L + + + N+ Y+ + YL D+QLF KRL + + +
Sbjct 93 ILGPADLTED----------ERTNLLNKFYLFGDVPYLRKTDLQLFLKRLRYYVTKQKPS 142
Query 180 EKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSWRFGDTDTQPVWSSASCY 239
EK+ Y+ VGEYGP FRPH+H+LLF S E Q + +SK+W FG D Q S Y
Sbjct 143 EKVRYFAVGEYGPVHFRPHYHLLLFLQSDEALQICSENISKAWTFGRVDCQVSKGQCSNY 202
Query 240 VAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKPQEDEEIFSLFYDGRM--- 296
VA YVNS+ +P +K S + PF S + F + E+I+SL + +
Sbjct 203 VASYVNSSCTIPKVFKA-SSVCPFSVHSQKLGQG-----FLDCQREKIYSLTPENFIRSS 256
Query 297 LELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATALSNIPHVLAKFGFIDEVTD 356
+ LNGK RS + YPR S + A S + A+ F D T
Sbjct 257 IVLNGKYKEFDVWRSCYSFFYPRCKGFVTKSSRE-----RAYSYSIYDTARLLFPDAKTT 311
Query 357 FEMSKRI 363
F ++K I
Sbjct 312 FSLAKEI 318
>gi|575094322|emb|CDL65709.1| unnamed protein product [uncultured bacterium]
Length=499
Score = 122 bits (305), Expect = 3e-27, Method: Compositional matrix adjust.
Identities = 100/358 (28%), Positives = 154/358 (43%), Gaps = 45/358 (13%)
Query 7 KAYGSCLHPRVIKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTY 66
+++ C P V+++ G P VPCG C C +NK LK ++ SKYC F+TLTY
Sbjct 3 QSFVKCFSPLVLRDP-RGYPYQVPCGKCIACHNNKRSSLSLKLRLEEYTSKYCYFLTLTY 61
Query 67 STEYLPVGEFYKGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYF--------DFDT 118
+ LP+ R P YS + RN SF ++ DF
Sbjct 62 DDDNLPLFSVGLDTCATEFVRIYP----YSERL-----RNDSFISDFCSDLHNFDNDFVD 112
Query 119 QLSWESAQLLQKKTHLHYTSFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNIRSVT 178
++ + S ++ ++ H + Y L L YRD+QLF KRL ++I
Sbjct 113 KMDYYSDYVINYESKYHKSCV----------YGHGLYALLYYRDIQLFLKRLRKHIYKYY 162
Query 179 NEKIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIR---------------QFVSKSWR 223
EKI +Y++GEYG + RPH+H LLF +S L Q+ +F+ W+
Sbjct 163 GEKIRFYIIGEYGTKSLRPHWHCLLFFNSSSLSQAFEDCVNVGTTSRPCSCPRFLRPFWQ 222
Query 224 FGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKPQE 283
FG D++ A YV+ YVN +A P S+ K + + S + V Q+
Sbjct 223 FGICDSKRTNGEAYNYVSSYVNQSANFPKLLVLLSNQKAYHSIQLGQILSEQSIVSAIQK 282
Query 284 DEEIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATALSNI 341
+ FS F L+ G RS+ +R +P+ S + + RV T +
Sbjct 283 GD--FSFFERQFYLDTFGAANSYSVWRSYYSRFFPKFTCSSQLTYEQTYRVLTCYETL 338
>gi|494822887|ref|WP_007558295.1| hypothetical protein [Bacteroides plebeius]
gi|198272100|gb|EDY96369.1| hypothetical protein BACPLE_00805 [Bacteroides plebeius DSM 17135]
Length=545
Score = 114 bits (286), Expect = 1e-24, Method: Compositional matrix adjust.
Identities = 115/422 (27%), Positives = 166/422 (39%), Gaps = 78/422 (18%)
Query 11 SCLHPRVIKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEY 70
SCL P+ IKNKYTG+ + V C C C + K C+ + +K F+TLT+ ++
Sbjct 7 SCLEPQRIKNKYTGEEMVVACKHCVACEQLRNFKYSNLCDFESLTAKKTVFLTLTFDDKF 66
Query 71 LPVGEFYKGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSWESAQLLQK 130
+P FYK E R D DT + L+
Sbjct 67 VPQFRFYKVGDDEYIMR---------------------------DADTG-EYLGRTLMTP 98
Query 131 KTHLHYTSFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNIRSVTNEKIYYYVVGEY 190
+ Y +R+ R + YL+ R++QLF KRL + + +KI ++ GEY
Sbjct 99 QLMNEYQ-----KRVNYRINYKGRFPYLSKRELQLFMKRLRKYLDKYEGQKIRFFATGEY 153
Query 191 GPTTFRPHFHILLFHDSKE-----------------------------LRQSIRQFVSKS 221
GP +FRPHFHILLF D L + ++ +S
Sbjct 154 GPLSFRPHFHILLFVDDPSLFLPSVHTLGEYPYPYWSKYQKAHCGKGTLLSKLEYYIRES 213
Query 222 WRFGDTDTQPV-WSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFK 280
W FG D Q V S S YVAGYVNS+ LP K +K F + S F
Sbjct 214 WPFGGIDAQSVEQGSCSSYVAGYVNSSVPLPSCLK-VDAVKSFSQHSRFLGRKIFGTELI 272
Query 281 PQEDEEIFSLFYDGRMLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATALSN 340
P + F+ F R G+ R ++ +YP+ S + RV T S
Sbjct 273 PLLKLK-FTEFVQ-RSFFCRGRYDNFRTPSEMLHSVYPQCKGFALLSHEQRFRVYTIWSR 330
Query 341 IPHVLAKFGFIDEVTDFEMSKRIYYLIRRYLEIDHTLKYAPEQLR----LIYNYLSFVVV 396
+ + D+ D S + Y +D + PE++R LIY LS +
Sbjct 331 LRYYFNS----DKKADVARS----LVTSFYSWLDTGILRVPERVREDFLLIYTELSQNLN 382
Query 397 YK 398
YK
Sbjct 383 YK 384
>gi|575094355|emb|CDL65737.1| unnamed protein product [uncultured bacterium]
Length=517
Score = 110 bits (276), Expect = 2e-23, Method: Compositional matrix adjust.
Identities = 96/339 (28%), Positives = 145/339 (43%), Gaps = 68/339 (20%)
Query 12 CLHPRVIKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYL 71
CL P+ + N Y D + VPCG C C +KA + +L+ ++ + K+C F TLTY+ Y+
Sbjct 11 CLEPKRVFNPYLNDWLLVPCGKCRACQCSKASRYKLQIQLEASQHKFCIFGTLTYANTYI 70
Query 72 PVGEFYKGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSWESAQLLQKK 131
P R +P + ++ V GY EY + S++ LL K
Sbjct 71 P------------RLSLVPYN-DKTFGVVNGYEMCDKETGEYLGYLDSPSYDVESLLDK- 116
Query 132 THLHYTSFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNIRSVTNEKIYYYVVGEYG 191
HL F D + YL RD+QLF KRL +N+ ++ K+ Y+ +GEYG
Sbjct 117 LHL----FGD-------------VPYLRKRDLQLFIKRLRKNLSKYSDAKVRYFAMGEYG 159
Query 192 PTTFRPHFHILLFHDS----------------------------KELRQSIRQFVSKSWR 223
P FRPH+H LLF D ++ + + SW+
Sbjct 160 PVHFRPHYHFLLFFDEIKFTAPSGHTLGEFPDWAWYDSQNKCSRSDILSVVEYCIRSSWK 219
Query 224 FGDTDTQPVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKPQE 283
FG D Q A+ YV+ YV+ + LP Y+ S +PF S + F E
Sbjct 220 FGRVDAQYSKGDAAQYVSSYVSGSGSLPKVYQ-VSSARPFSLHSRFLGQG-----FLAHE 273
Query 284 DEEIFSL---FYDGRMLELNGKPTLVRPKRSHINRLYPR 319
E+++ + R +ELNG RS + YP+
Sbjct 274 CEKVYETPVRDFVKRSVELNGSNKDFNLWRSCYSVFYPK 312
>gi|494610270|ref|WP_007368516.1| hypothetical protein [Prevotella multiformis]
gi|324988542|gb|EGC20505.1| hypothetical protein HMPREF9141_0984 [Prevotella multiformis
DSM 16608]
Length=479
Score = 106 bits (265), Expect = 5e-22, Method: Compositional matrix adjust.
Identities = 95/345 (28%), Positives = 152/345 (44%), Gaps = 66/345 (19%)
Query 10 GSCLHPRVIKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTE 69
SCL P+ I NKY + +YVPC C C + A + + ++ F+TLTY E
Sbjct 8 NSCLSPKRIYNKYIDETLYVPCRKCFRCRDSYASDWSRRIENECREHRFSLFVTLTYDNE 67
Query 70 YLPVGEFYKGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSWESAQLLQ 129
++P+ F D + W S +L +
Sbjct 68 HIPL-----------------------------------FQPLVMDDGSHPVWFSNRLSE 92
Query 130 KKTHLHYT---SFPDGRRIYNRPYMENLI--GYLNYRDMQLFFKRLNQNI-----RSVTN 179
L + S P + ME+ + Y +D+Q +FKRL + ++ +N
Sbjct 93 SGKFLSDSVCRSLPPQK-------MEDEVCFAYPCKKDVQDWFKRLRSAVDYQLNKNKSN 145
Query 180 E-KIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSWRFGDTDTQPVWSSASC 238
E +I Y++ EYGP TFRPH+H +L++DS+EL+++I + + ++W+ G++ V +SAS
Sbjct 146 EFRIRYFICSEYGPRTFRPHYHAILWYDSEELQRNIGRLIRETWKNGNSVFSLVNNSASQ 205
Query 239 YVAGYVNSTACLPDFYKN-FSHIKPFGRFSMHFAESAFNEVFKPQEDEEIFSLFYDGR-- 295
YVA YVN LP F + F+ + H A + ++E + S DG
Sbjct 206 YVAKYVNGDTRLPPFLRTEFTS-------TFHLASKHPYIGYCKADEEALRSNVLDGTYG 258
Query 296 ---MLELNGKPTLVRPKRSHINRLYPRLNKSKHASVDDDIRVATA 337
+ NG+ V RS NRL P+ + S + IRV A
Sbjct 259 QSVLNRDNGQFEFVPTPRSLENRLLPKCRGYRSLSHSERIRVYAA 303
>gi|565841285|ref|WP_023924566.1| hypothetical protein [Prevotella nigrescens]
gi|564729906|gb|ETD29850.1| hypothetical protein HMPREF1173_00032 [Prevotella nigrescens
CC14M]
Length=484
Score = 105 bits (262), Expect = 1e-21, Method: Compositional matrix adjust.
Identities = 72/252 (29%), Positives = 113/252 (45%), Gaps = 47/252 (19%)
Query 12 CLHPRVIKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYL 71
C HP+ I N YT + V+V C C+ C++ K + + Y F+TLTY E+L
Sbjct 9 CEHPKRIINPYTHERVWVACRRCKCCLNKKTSAWSGRVANECKLHAYSAFVTLTYDNEHL 68
Query 72 PVGEFYKGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSWESAQLLQKK 131
P+ + E + ++ W S +L +K
Sbjct 69 PL-----------------------------------YQPECMNERGEMVWTSNRLCDEK 93
Query 132 THLHYTSFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNI-------RSVTNEKIYY 184
+ F ++ N + Y D+ FFKRL + +TNEKI Y
Sbjct 94 VIVGNYDFI---KVSNSDVQA--VAYCCKSDIVKFFKRLRSKLSYYFKKHHIITNEKIRY 148
Query 185 YVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSWRFGDTDTQPVWSSASCYVAGYV 244
+V EYGP T RPH+H +++ DS+E+ + I + +S SW G TD + V S+A YVA YV
Sbjct 149 FVCSEYGPKTLRPHYHAIIWFDSEEVARVIEKMLSSSWSNGFTDFEYVNSTAPQYVAKYV 208
Query 245 NSTACLPDFYKN 256
+ + LP+ ++
Sbjct 209 SGNSVLPEILQH 220
>gi|575094298|emb|CDL65688.1| unnamed protein product [uncultured bacterium]
Length=478
Score = 100 bits (250), Expect = 5e-20, Method: Compositional matrix adjust.
Identities = 94/334 (28%), Positives = 143/334 (43%), Gaps = 46/334 (14%)
Query 12 CLHPRVIKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEYL 71
C++ R I+NKYTG +YV CG C C+ KA + K ++ C F+TL Y ++
Sbjct 2 CINKREIRNKYTGQKLYVSCGKCPACLQEKANASAYKIRNNQSSELSCFFVTLNYDNNHI 61
Query 72 PV---GEFYKGASGEV-------RFRCLPRDFVYSYKTVQGYNR----NISFNDEYFDFD 117
PV + Y S +V + CLP D +Y N+ N N D
Sbjct 62 PVIFKHDVYNYNSSDVYHFDEERKELCLPVD-LYRGVCPAFSNKIDTFNFPLNRLSTDVV 120
Query 118 TQLSWESAQLLQKKTHLHYTSFPDGRRIYNRPYM--ENLIGYLNYRDMQLFFKRLNQNIR 175
+ L +++ K H +P + E + +D+QLFFKRL Q++
Sbjct 121 SSLDNHCGVVVKTKNH--------------KPVLFNEEIFSVCYTKDIQLFFKRLRQSLY 166
Query 176 SVTNEK--IYYYVVGEYGPTTFRPHFHILLFHDSKELR-QSIRQFVSKSWRFGDTDTQ-- 230
+ I Y+ EYGPTT+R HFH+ +F E+ S R+ K+W F
Sbjct 167 RKFGFRPFIQYFQTSEYGPTTYRAHFHLCIFVKRSEISFDSFRKACVKAWPFCSKKQMFR 226
Query 231 --PVWSSASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAES----AFNEVFKPQED 284
+ S S Y+A YVN A +P F N K S++F + +FN + E
Sbjct 227 NVEIARSPSAYIASYVNCRANVPLFL-NLKEAKAKHTHSLYFGHNNDKLSFNSIVNRFET 285
Query 285 EEIFSLFYDGRMLELNGKP-TLVRPKRSHINRLY 317
+ + Y + ++G P T P +IN Y
Sbjct 286 QG--TTLYPRELSSVDGVPQTSFLPLPRYINAYY 317
>gi|517172763|ref|WP_018361581.1| hypothetical protein [Prevotella nanceiensis]
Length=598
Score = 99.8 bits (247), Expect = 2e-19, Method: Compositional matrix adjust.
Identities = 65/219 (30%), Positives = 103/219 (47%), Gaps = 31/219 (14%)
Query 11 SCLHPRVIKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFITLTYSTEY 70
CL P I NK TG VPC C +C++ +A K + ++ + TLTY Y
Sbjct 25 GCLSPSYIYNKNTGHYETVPCHNCTYCVNVEASKQSRRVREEIKQHLFSVMFTLTYDNVY 84
Query 71 LPVGEFYKGASGEVRFRCLPR--DFVYSYK-TVQGYNRNISFNDEYFDFDTQLSWESAQL 127
+P E + G GE++ + + R D S + YN + FND DT++ W
Sbjct 85 IPRMEAFAGKHGEMQLKPIGRTADLHDSCPFNSKNYNGDYRFND-----DTRIPWIENNK 139
Query 128 LQKKTHLHYTSFPDGRRIYNRPYMENLIGYLNYRDMQLFFKRLNQNIRSV----TNEKIY 183
+ K +L + + ++ +D+Q F KRL + I + +KI
Sbjct 140 IYCKNNLQFAT-------------------VSKKDIQNFLKRLRKKIDKLNIPQNEKKIR 180
Query 184 YYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSW 222
Y++ EYGP T+RPH+H +LF DS + I+ F+ +SW
Sbjct 181 YFIASEYGPKTYRPHYHGVLFIDSPTVLSKIKAFIVESW 219
>gi|647452984|ref|WP_025792805.1| hypothetical protein [Prevotella histicola]
Length=480
Score = 97.8 bits (242), Expect = 6e-19, Method: Compositional matrix adjust.
Identities = 92/348 (26%), Positives = 151/348 (43%), Gaps = 67/348 (19%)
Query 6 SKAYGS---CLHPRVIKNKYTGDPVYVPCGTCEFCIHNKAIKAELKCNVQLAASKYCEFI 62
S+A G+ CL P I N+Y G+ +Y C C C + A + + + + +Y F+
Sbjct 3 SRALGNINPCLRPHRIYNRYIGEFLYTNCRKCVRCRSSYASSWANRIDSECSFHRYSLFL 62
Query 63 TLTYSTEYLPVGEFYKGASGEVRFRCLPRDFVYSYKTVQGYNRNISFNDEYFDFDTQLSW 122
TLTY ++LP + + G ++ ++ D +S
Sbjct 63 TLTYDNDHLP--------------------YYAPLFNLDGSRTDVWCSNRGCDNGKFVSS 102
Query 123 ESAQLLQKKTHLHYTSFPDGRRIYNRPYMENLI--GYLNYRDMQLFFKRLNQNI----RS 176
+ A+ + P G ME+ + Y +D+Q FFKRL I +
Sbjct 103 DIARPIP----------PVG--------MEDTVCFAYPCKKDVQDFFKRLRSKIDYKLKP 144
Query 177 VTNE-KIYYYVVGEYGPTTFRPHFHILLFHDSKELRQSIRQFVSKSWRFGDTDTQPVWSS 235
NE +I Y++ EYGP TFRPH+H +L++DS+ L + + ++W+ G+TD V SS
Sbjct 145 RGNEYRIRYFICSEYGPNTFRPHYHAILWYDSEILHNELNVLIRETWKNGNTDFSLVNSS 204
Query 236 ASCYVAGYVNSTACLPDFYKNFSHIKPFGRFSMHFAESAFNEVFKPQEDEEIFSLFYDGR 295
AS YVA YVN LP F + F+ F ++ + +D+E Y+
Sbjct 205 ASQYVAKYVNGDCDLPSFLRT--------EFTSTFHLASKHPCIGYGKDDE--EALYENV 254
Query 296 MLELNGKPTL---------VRPKRSHINRLYPRLNKSKHASVDDDIRV 334
+ G+ L V P RS NR+ P+ + S + +R+
Sbjct 255 INGTYGRNCLNKSTNEFEFVCPPRSLENRILPKCKGYRRISHSERVRI 302
Lambda K H a alpha
0.325 0.140 0.435 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 2552904812250