bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-19_CDS_annotation_glimmer3.pl_2_1
Length=75
Score E
Sequences producing significant alignments: (Bits) Value
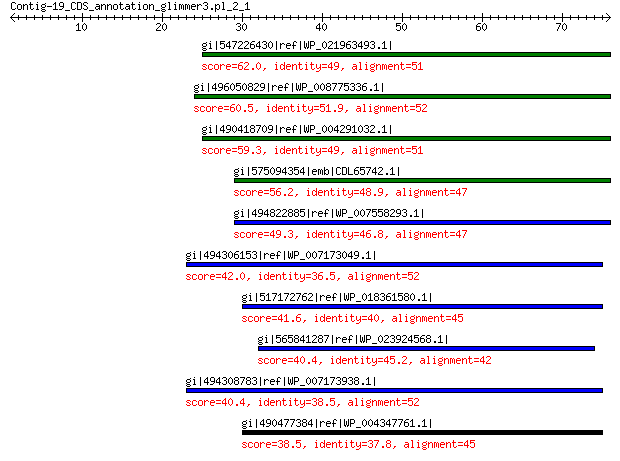
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 62.0 3e-09
gi|496050829|ref|WP_008775336.1| hypothetical protein 60.5 1e-08
gi|490418709|ref|WP_004291032.1| hypothetical protein 59.3 3e-08
gi|575094354|emb|CDL65742.1| unnamed protein product 56.2 3e-07
gi|494822885|ref|WP_007558293.1| hypothetical protein 49.3 6e-05
gi|494306153|ref|WP_007173049.1| hypothetical protein 42.0 0.016
gi|517172762|ref|WP_018361580.1| hypothetical protein 41.6 0.022
gi|565841287|ref|WP_023924568.1| hypothetical protein 40.4 0.059
gi|494308783|ref|WP_007173938.1| hypothetical protein 40.4 0.066
gi|490477384|ref|WP_004347761.1| capsid protein 38.5 0.26
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 62.0 bits (149), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 25/51 (49%), Positives = 37/51 (73%), Gaps = 0/51 (0%)
Query 25 LSYTWFKVNPSVLNPIFGVAVDGSWNSDQLLCNCQFNVKVARNLSYDGMPY 75
++Y +FKVNP +++ IFGV D + N+DQLL N F++K RN Y+G+PY
Sbjct 523 MTYNFFKVNPHIVDNIFGVKADSTINTDQLLINSYFDIKAVRNFDYNGLPY 573
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 60.5 bits (145), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 27/52 (52%), Positives = 39/52 (75%), Gaps = 0/52 (0%)
Query 24 ILSYTWFKVNPSVLNPIFGVAVDGSWNSDQLLCNCQFNVKVARNLSYDGMPY 75
+++YT FKVNP+ ++P+F VA S ++DQ LC+ F+VKV RNL DG+PY
Sbjct 529 LVNYTNFKVNPNCVDPLFAVAASNSIDTDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 59.3 bits (142), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 25/51 (49%), Positives = 35/51 (69%), Gaps = 0/51 (0%)
Query 25 LSYTWFKVNPSVLNPIFGVAVDGSWNSDQLLCNCQFNVKVARNLSYDGMPY 75
+++T+FKVNP L+PIF V N+DQ LC+ F++K RNL DG+PY
Sbjct 528 MNFTFFKVNPDCLDPIFAVQAGDDTNTDQFLCSSFFDIKAVRNLDTDGLPY 578
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 56.2 bits (134), Expect = 3e-07, Method: Compositional matrix adjust.
Identities = 23/47 (49%), Positives = 35/47 (74%), Gaps = 0/47 (0%)
Query 29 WFKVNPSVLNPIFGVAVDGSWNSDQLLCNCQFNVKVARNLSYDGMPY 75
+FKVNPS+++P+F V D + +D+ LC+ F+VKV RNL +G+PY
Sbjct 569 FFKVNPSIVDPLFAVVADSTVKTDEFLCSSFFDVKVVRNLDVNGLPY 615
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 49.3 bits (116), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 22/47 (47%), Positives = 33/47 (70%), Gaps = 0/47 (0%)
Query 29 WFKVNPSVLNPIFGVAVDGSWNSDQLLCNCQFNVKVARNLSYDGMPY 75
+FKV+PSVL+ +F V + N+DQ LC+ F+V V R+L +G+PY
Sbjct 567 FFKVSPSVLDNLFAVKANSDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|494306153|ref|WP_007173049.1| hypothetical protein [Prevotella bergensis]
gi|270333881|gb|EFA44667.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=519
Score = 42.0 bits (97), Expect = 0.016, Method: Compositional matrix adjust.
Identities = 19/52 (37%), Positives = 30/52 (58%), Gaps = 0/52 (0%)
Query 23 PILSYTWFKVNPSVLNPIFGVAVDGSWNSDQLLCNCQFNVKVARNLSYDGMP 74
P L FK++P LN +F V +G+ ++D + C FN+ ++S DGMP
Sbjct 466 PQLEIADFKIDPGCLNSVFPVEFNGTESTDCVFGGCNFNIVKVSDMSVDGMP 517
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 41.6 bits (96), Expect = 0.022, Method: Compositional matrix adjust.
Identities = 18/45 (40%), Positives = 28/45 (62%), Gaps = 0/45 (0%)
Query 30 FKVNPSVLNPIFGVAVDGSWNSDQLLCNCQFNVKVARNLSYDGMP 74
FK+NP L+ +F V +G+ +DQ+ C FN+ ++S DGMP
Sbjct 522 FKINPKWLDDVFAVNYNGTELTDQVFGGCYFNIVKVSDMSIDGMP 566
>gi|565841287|ref|WP_023924568.1| hypothetical protein [Prevotella nigrescens]
gi|564729907|gb|ETD29851.1| hypothetical protein HMPREF1173_00033 [Prevotella nigrescens
CC14M]
Length=656
Score = 40.4 bits (93), Expect = 0.059, Method: Compositional matrix adjust.
Identities = 19/42 (45%), Positives = 26/42 (62%), Gaps = 0/42 (0%)
Query 32 VNPSVLNPIFGVAVDGSWNSDQLLCNCQFNVKVARNLSYDGM 73
V+P VL PIF V +GS ++DQ L N F+VK R + + M
Sbjct 612 VDPKVLEPIFAVKYNGSMSTDQFLVNSYFDVKAIRPMQVNDM 653
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 40.4 bits (93), Expect = 0.066, Method: Compositional matrix adjust.
Identities = 20/52 (38%), Positives = 29/52 (56%), Gaps = 0/52 (0%)
Query 23 PILSYTWFKVNPSVLNPIFGVAVDGSWNSDQLLCNCQFNVKVARNLSYDGMP 74
P L FK++P LN IF V +G+ +D + C FN+ ++S DGMP
Sbjct 500 PQLEIADFKIDPGCLNSIFPVDYNGTEANDCVYGGCNFNIVKVSDMSVDGMP 551
>gi|490477384|ref|WP_004347761.1| capsid protein [Prevotella buccalis]
gi|281300712|gb|EFA93043.1| putative capsid protein (F protein) [Prevotella buccalis ATCC
35310]
Length=552
Score = 38.5 bits (88), Expect = 0.26, Method: Composition-based stats.
Identities = 17/45 (38%), Positives = 27/45 (60%), Gaps = 0/45 (0%)
Query 30 FKVNPSVLNPIFGVAVDGSWNSDQLLCNCQFNVKVARNLSYDGMP 74
K+NP L+ IF V +G+ +D + CQFNV+ ++S +G P
Sbjct 506 LKINPKWLDSIFAVNYNGTQITDCVFGGCQFNVQKVSDMSENGEP 550
Lambda K H a alpha
0.321 0.141 0.466 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 443954776629