bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-18_CDS_annotation_glimmer3.pl_2_6
Length=303
Score E
Sequences producing significant alignments: (Bits) Value
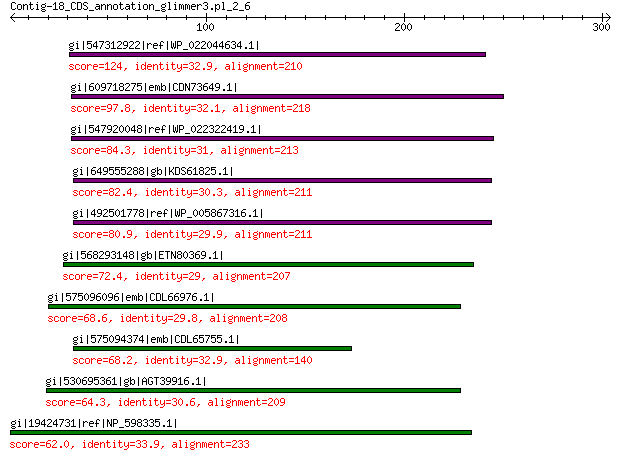
gi|547312922|ref|WP_022044634.1| putative replication initiation... 124 1e-29
gi|609718275|emb|CDN73649.1| conserved hypothetical protein 97.8 2e-20
gi|547920048|ref|WP_022322419.1| putative replication protein 84.3 8e-16
gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 82.4 4e-15
gi|492501778|ref|WP_005867316.1| hypothetical protein 80.9 2e-14
gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 72.4 3e-11
gi|575096096|emb|CDL66976.1| unnamed protein product 68.6 5e-10
gi|575094374|emb|CDL65755.1| unnamed protein product 68.2 1e-09
gi|530695361|gb|AGT39916.1| replication initiator 64.3 2e-08
gi|19424731|ref|NP_598335.1| hypothetical protein Sp-4p1 62.0 1e-07
>gi|547312922|ref|WP_022044634.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
gi|524208442|emb|CCZ76638.1| putative replication initiation protein [Alistipes finegoldii
CAG:68]
Length=320
Score = 124 bits (312), Expect = 1e-29, Method: Compositional matrix adjust.
Identities = 69/227 (30%), Positives = 128/227 (56%), Gaps = 31/227 (14%)
Query 31 IQIPCGHCEECRRAKANEWRVRLMEEIKSYPKN-IIFATLTFSEESLKKLEYDEKEPNKA 89
+++PCG+C C+++ N++R+RL+ E++ YP +F TLTF+++SL+K D
Sbjct 40 LEVPCGYCHSCQKSYNNQYRIRLLYELRKYPPGTCLFVTLTFNDDSLEKFSKDT------ 93
Query 90 PQKAISLFRKRWWKKYKTPLKHWLITEMGHDNTKRIHLHGIIWT--ELTEEQFEKE---- 143
KA+ LF R+ K Y ++HW + E G + R H HGI++ + + ++ +
Sbjct 94 -NKAVRLFLDRFRKVYGKQIRHWFVCEFGTLH-GRPHYHGILFNVPQALIDGYDSDMPGH 151
Query 144 -------WGYGWIFFGYEVSEKTINYIVKYVTKR---DEANPEFNGKIFTSKGIGIGYIN 193
W YG++F GY VS++T +YI KYVTK D+ P ++ +S GIG Y+N
Sbjct 152 HPLLASCWKYGFVFVGY-VSDETCSYITKYVTKSINGDKVRP----RVISSFGIGSNYLN 206
Query 194 KNSLNKHRYQDKFTEETYRAASGIKLALPTYYKQKLWTDQERESLRI 240
+ H+ ++ + + +G + A+P YY K+++D +++++ +
Sbjct 207 TEESSLHKLGNQ-RYQPFMVLNGFQQAMPRYYYNKIFSDVDKQNMVV 252
>gi|609718275|emb|CDN73649.1| conserved hypothetical protein [Elizabethkingia anophelis]
Length=265
Score = 97.8 bits (242), Expect = 2e-20, Method: Compositional matrix adjust.
Identities = 70/226 (31%), Positives = 112/226 (50%), Gaps = 17/226 (8%)
Query 32 QIPCGHCEECRRAKANEWRVRLMEEIKSYPKNIIFATLTFSEESLKKLEYDEKEPNKAPQ 91
PCG C ECR+A+ N W RL EE+K K+ F TLT+S+ L Y +
Sbjct 22 SFPCGKCLECRKARTNSWFARLTEELKV-SKSAHFVTLTYSDV---YLPYSDNGLISLDY 77
Query 92 KAISLFRKRWWKKYKTPLKHWLITEMGHDNTKRIHLHGIIWTELTEEQFEKEWGYGWIFF 151
+ LF KR K K+ +K++L+ E G T R H H I++ + F EW G +
Sbjct 78 RDFQLFMKRARKLQKSKIKYFLVGEYGA-QTYRPHYHAIVFGVENIDAFLGEWRMGNVHA 136
Query 152 GYEVSEKTINYIVKYVTKRDEANPEFNG-------KIFTSKGIGIGYINKNSLNKHRYQD 204
G V+ K+I Y +KY TK P+ + K SKG+G+ ++ ++ + +Y
Sbjct 137 G-TVTAKSIYYTLKYCTKSITEGPDKDPDDDRKPEKALMSKGLGLSHLTESMI---KYYK 192
Query 205 KFTEETYRAASGIKLALPTYYKQKLWTDQER-ESLRIIKEEQQVKY 249
++ G +ALP YY+ K+++D E+ L I + +++Y
Sbjct 193 DDVSRSFSLLGGTTIALPRYYRDKVFSDIEKVHRLVSITDYLEIRY 238
>gi|547920048|ref|WP_022322419.1| putative replication protein [Parabacteroides merdae CAG:48]
gi|524592960|emb|CDD13572.1| putative replication protein [Parabacteroides merdae CAG:48]
Length=278
Score = 84.3 bits (207), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 66/231 (29%), Positives = 118/231 (51%), Gaps = 24/231 (10%)
Query 32 QIPCGHCEECRRAKANEWRVRLMEEIKSYPKNIIFATLTFSEESL--KKLEYDEKEPNKA 89
++PCG C CR+ K W RL E K YP + +F TLT+ +E L +++ D + N A
Sbjct 9 KVPCGWCVNCRQNKRQSWVYRLQAEAKEYPLS-LFVTLTYDDEHLPIERIGSDLFQTNVA 67
Query 90 --PQKAISLFRKRWWKKYKT-PLKHWLITEMGHDNTKRIHLHGIIW-----TELTEEQFE 141
++ + LF KR KKY+ +++++ +E G N R H H I++ ++ +
Sbjct 68 VVSKRDVQLFMKRLRKKYEDYKMRYFVTSEYGAKNG-RPHYHMILFGFPFTGKMAGDLLA 126
Query 142 KEWGYGWIFFGYEVSEKTINYIVKYVTK--------RDEANPEFNGKIFTSKGIGIGYIN 193
+ W G++ + ++ K I Y+ KY+ + RDE + + GIG G++
Sbjct 127 ECWQNGFV-QAHPLTIKEIAYVCKYMYEKSMCPEILRDEKKYKPFMLCSRNPGIGFGFMK 185
Query 194 KNSLNKHRYQDKFTEETYRAASGIKLALPTYYKQKLWTDQERESLRIIKEE 244
+ + +R + + RA +G K+A+P YY KL+ D + L+ ++EE
Sbjct 186 ADIIEFYR---RHPRDYVRAWAGHKMAMPRYYADKLYDDDMKAFLKEMREE 233
>gi|649555288|gb|KDS61825.1| hypothetical protein M095_3808 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649560564|gb|KDS66872.1| hypothetical protein M095_2449 [Parabacteroides distasonis str.
3999B T(B) 4]
gi|649561011|gb|KDS67298.1| hypothetical protein M095_2409 [Parabacteroides distasonis str.
3999B T(B) 4]
Length=284
Score = 82.4 bits (202), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 64/229 (28%), Positives = 112/229 (49%), Gaps = 24/229 (10%)
Query 33 IPCGHCEECRRAKANEWRVRLMEEIKSYPKNIIFATLTFSEESLKKLEYDEK----EPNK 88
+PCG C CR+ K W RL E YP + +F TLT+ +E + E
Sbjct 15 VPCGRCVNCRKNKRQSWVYRLQAEADEYPFS-LFVTLTYDDEHIPTAMIGEDLFKTTVGV 73
Query 89 APQKAISLFRKRWWKKY-KTPLKHWLITEMGHDNTKRIHLHGIIW-----TELTEEQFEK 142
++ I LF KR KKY + L+++L +E G R H H I++ + + +
Sbjct 74 VSKRDIQLFMKRLRKKYAQYRLRYFLTSEYG-SQGGRPHYHMILFGFPFTGKHGGDLLAE 132
Query 143 EWGYGWIFFGYEVSEKTINYIVKYVTKRD------EANPEFNGKIFTSKGIGIGY--INK 194
W G++ + ++ K I+Y+ KY+ ++ + E+ + SK GIGY + +
Sbjct 133 CWKNGFV-QAHPLTTKEISYVTKYMYEKSMIPDILKGVKEYQPFMLCSKMPGIGYHFLRE 191
Query 195 NSLNKHRYQDKFTEETYRAASGIKLALPTYYKQKLWTDQERESLRIIKE 243
L+ +R + + RA +G+++A+P YY KL+ D +E L+ ++E
Sbjct 192 QILDFYRLHPR---DYVRAFNGMRMAMPRYYADKLYDDDMKEYLKELRE 237
>gi|492501778|ref|WP_005867316.1| hypothetical protein [Parabacteroides distasonis]
gi|409230407|gb|EKN23271.1| hypothetical protein HMPREF1059_03256 [Parabacteroides distasonis
CL09T03C24]
Length=284
Score = 80.9 bits (198), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 63/229 (28%), Positives = 110/229 (48%), Gaps = 24/229 (10%)
Query 33 IPCGHCEECRRAKANEWRVRLMEEIKSYPKNIIFATLTFSEESLKKLEYDE----KEPNK 88
+PCG C CR+ K W RL E YP + +F TLT+ +E + E
Sbjct 15 VPCGRCVNCRKNKRQSWVYRLQAEADEYPFS-LFVTLTYDDEHMPTAMIGEDLFKSTVGV 73
Query 89 APQKAISLFRKRWWKKY-KTPLKHWLITEMGHDNTKRIHLHGIIW-----TELTEEQFEK 142
++ I LF KR KKY + L+++L +E G R H H I++ + + +
Sbjct 74 VSKRDIQLFMKRLRKKYDQYRLRYFLTSEYG-SQGGRPHYHMILFGFPFTGKHGGDLLAE 132
Query 143 EWGYGWIFFGYEVSEKTINYIVKYVTKRDEANP------EFNGKIFTSKGIGIGY--INK 194
W G++ + ++ K I Y+ KY+ ++ E+ + S+ GIGY + +
Sbjct 133 CWKNGFV-QAHPLTTKEIAYVTKYMYEKSMVPDILKDVKEYQPFMLCSRIPGIGYHFLRE 191
Query 195 NSLNKHRYQDKFTEETYRAASGIKLALPTYYKQKLWTDQERESLRIIKE 243
L+ +R + + RA +G+++A+P YY KL+ D +E L+ ++E
Sbjct 192 QILDFYRLHPR---DYVRAFNGMRMAMPRYYADKLYDDDMKEYLKELRE 237
>gi|568293148|gb|ETN80369.1| hypothetical protein NECAME_18023 [Necator americanus]
Length=345
Score = 72.4 bits (176), Expect = 3e-11, Method: Compositional matrix adjust.
Identities = 60/227 (26%), Positives = 102/227 (45%), Gaps = 37/227 (16%)
Query 28 LRWIQIPCGHCEECRRAKANEWRVRLMEEIKSYPKNIIFATLTFSEESLKKLEYDEKEPN 87
L + +PCG C C+R + + W RL++E + +N F TLT YD +
Sbjct 15 LEKVPVPCGRCPPCKRRRVDSWVFRLLQEELQH-ENASFVTLT----------YDTRFVP 63
Query 88 KAPQKAISLFR---KRWWKKYK--TP---LKHWLITEMGHDNTKRIHLHGIIWTELTEEQ 139
+ ++L R R+ K+ + P LK+++ E G R H H II+ +
Sbjct 64 ISKNGFMTLDRGEFPRYMKRLRKLVPGRKLKYYMCGEYGSQRF-RPHYHAIIFGVPQDSL 122
Query 140 FEKEWGYGWIFF----GYEVSEKTINYIVKYVTK--------RDEANPEFNGKIFTSKGI 187
F W V+ K+I Y +KY+ K RD+ PEF+ SKG+
Sbjct 123 FADAWTLNGDSLGGVVVGTVTGKSIAYTMKYIDKSTWKQKHGRDDRVPEFS---LMSKGM 179
Query 188 GIGYINKNSLNKHRYQDKFTEETYRAASGIKLALPTYYKQKLWTDQE 234
G+ Y+ + H+ + + G ++A+P YY+QK+++D +
Sbjct 180 GVSYLTPQMVEYHK--EDISRLFCTREGGSRIAMPRYYRQKIYSDDD 224
>gi|575096096|emb|CDL66976.1| unnamed protein product [uncultured bacterium]
Length=296
Score = 68.6 bits (166), Expect = 5e-10, Method: Compositional matrix adjust.
Identities = 62/244 (25%), Positives = 103/244 (42%), Gaps = 56/244 (23%)
Query 20 GKRARDRRLRWIQIPCGHCEECRRAKANEWRVRLMEEIKSYPKNIIFATLTFSEESLKKL 79
G++ ++ ++ +PCG C ECR +A+EW +R E+KS+ K IF TLT+++++L
Sbjct 23 GRKEQEFSHDYVLVPCGQCLECRLHRASEWALRCCHELKSHDKG-IFLTLTYNDDNL--- 78
Query 80 EYDEKEPNKA-PQKAISLFRKRWWKKYK-----TPLKHWLITEMGHDNTKRIHLHGIIW- 132
PN +K + F KR + T +++ E G D + R H H +++
Sbjct 79 -----PPNGTLVKKHVQDFIKRLRRHIDYYGDCTKIRYLCAGEYG-DLSLRPHYHLLVFG 132
Query 133 ------------------TELTEEQFEKEWGYGWIFFGYEVSEKTINYIVKYVTKRDEAN 174
+ T K WG G I FG ++ ++ Y +Y K+
Sbjct 133 YYPSDPRLLHGLQKIGKNSLFTSPTLTKLWGKGHISFG-AITFESARYTCQYALKKQTGE 191
Query 175 -----------PEFNGKIFTSKGIGIGYINKNSLNKHRYQDKFTEETYRAASGIKLALPT 223
PEF + S G+GY S D E Y +G K+ +P
Sbjct 192 HSHYYVDRGVIPEF---MICSNRNGLGYDFAVS------HDNMFERGYLTMNGKKIGIPR 242
Query 224 YYKQ 227
YY++
Sbjct 243 YYQK 246
>gi|575094374|emb|CDL65755.1| unnamed protein product [uncultured bacterium]
Length=487
Score = 68.2 bits (165), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 46/158 (29%), Positives = 76/158 (48%), Gaps = 23/158 (15%)
Query 33 IPCGHCEECRRAKANEWRVRLMEEIKSYPKNIIFATLTFSEESLKKLEYDEKEPNKAPQ- 91
+PCGHC +C+ AK +W+VR EE+ + ++ F TLT + P+ +P+
Sbjct 26 VPCGHCYDCKSAKTTDWQVRCSEELNNNSQS-YFYTLTLDPRFIDTY---GTLPDGSPRY 81
Query 92 ----KAISLFRKRWWK---KYKTPLKHWLITEMGHDNTKRIHLHGIIWTELTEEQFE--- 141
+ I LF KR K KY LK+ ++ E+G + T R H H I + + F+
Sbjct 82 VFNKRHIQLFLKRLRKALSKYNISLKYVIVGELG-ETTHRPHYHAIFYLSSSVNPFKFRI 140
Query 142 ---KEWGYGWIFFGYE----VSEKTINYIVKYVTKRDE 172
W G+I G ++ ++Y++KY+ K D
Sbjct 141 MVRNSWSLGFIKSGDNNGIILNNDAVSYVIKYMHKTDS 178
>gi|530695361|gb|AGT39916.1| replication initiator [Marine gokushovirus]
Length=289
Score = 64.3 bits (155), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 64/249 (26%), Positives = 95/249 (38%), Gaps = 54/249 (22%)
Query 19 NGKRARDRRL---RWIQIPCGHCEECRRAKANEWRVRLMEEIKSYPKNIIFATLTFSEES 75
+GK D+ R +PCG C CR + +W +R + E +++ N F TLTF E
Sbjct 13 DGKVVFDKPFAFARGFNLPCGQCIGCRLDYSRQWAIRCVHEAQTHEDN-CFITLTFDNEH 71
Query 76 LKKLEYDEKEPNKAPQKAISLFRKRWWKKYKTPLKHWLITEMGHDNTKRIHLHGIIWTE- 134
+ K K P F KR KKY ++ + E G D KR H H +++
Sbjct 72 IAK----RKNPESLDNTEFQRFMKRLRKKYPHKIRFFHCGEYG-DQNKRPHYHALLFGHD 126
Query 135 ---------------LTEEQFEKEWGYGWIFFGYEVSEKTINYIVKYVTKR--------- 170
++ + W YG+ G VS T Y +YV K+
Sbjct 127 FKDKKLWSNKGDFKLFVSQELAELWPYGFHTIG-AVSFDTAAYCARYVMKKVTGDAAASH 185
Query 171 ------------DEANPEFNGKIFTSKGIGIGYINKNSLNKHRYQDKFTEETYRAASGIK 218
+E PE+ S+ GIGY K+ Y D + Y +G K
Sbjct 186 YREVDLETGEVINEIKPEY---CTMSRMPGIGY---EWYQKYGYHDCHKHD-YIVINGYK 238
Query 219 LALPTYYKQ 227
+ P YY +
Sbjct 239 VRPPRYYDK 247
>gi|19424731|ref|NP_598335.1| hypothetical protein Sp-4p1 [Spiroplasma phage 4]
gi|137995|sp|P11334.1|REP_SPV4 RecName: Full=Replication-associated protein ORF2; AltName: Full=Rep
[Spiroplasma phage 4]
Length=320
Score = 62.0 bits (149), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 79/278 (28%), Positives = 118/278 (42%), Gaps = 60/278 (22%)
Query 1 VCLYPSIIENPKYA-----KSNKNGKRAR-DRRLRWI---QIPCGHCEECRRAKANEWRV 51
CL P + N K K NG AR D +I +PC C CR + EW V
Sbjct 2 ACLRPLQVHNLKKGEKVNFKHYSNGDVARYDMNKNYIVNDSVPCRKCVGCRLDNSAEWGV 61
Query 52 RLMEEIKSYPKNIIFATLTFSEESLKKLEYDEKEPNKAPQ---KAISLFRKRWWKKYKTP 108
R EIKS PK+ F TLT+S+E L + PN P+ K I RK + ++
Sbjct 62 RASLEIKSNPKHNWFVTLTYSDEHL--VYNALGRPNCVPEHITKFIKSLRKYFERRGHIG 119
Query 109 LKHWLITEMGHDNTKRIHLH-GIIWTELTEEQFEKE------------------WGYGWI 149
+K+ E G TKR+ H I + L + EK W G+
Sbjct 120 IKYLASNEYG---TKRMRPHYHICFFNLPLDDLEKTIDSQKGYQQWTSKTISRFWDKGFH 176
Query 150 FFGYEVSEKTINYIVKYVTKR--------DEANPEFNGKIFTSKGIGIGYINKNSLNKHR 201
G E++ + NY +Y TK+ + PE K+ SKGIG+ Y +N
Sbjct 177 TIG-ELTYHSANYTARYTTKKLGVKDYKALQLVPE---KLRMSKGIGLKYFMENK----- 227
Query 202 YQDKFTEETYRAAS--GIK-LALPTYYKQKL---WTDQ 233
+ + E++ ++ GIK +P Y+ +++ W D+
Sbjct 228 -ERIYKEDSVLISTDKGIKRFKVPKYFDRRMEREWQDE 264
Lambda K H a alpha
0.317 0.134 0.414 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1615949850528