bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-16_CDS_annotation_glimmer3.pl_2_2
Length=545
Score E
Sequences producing significant alignments: (Bits) Value
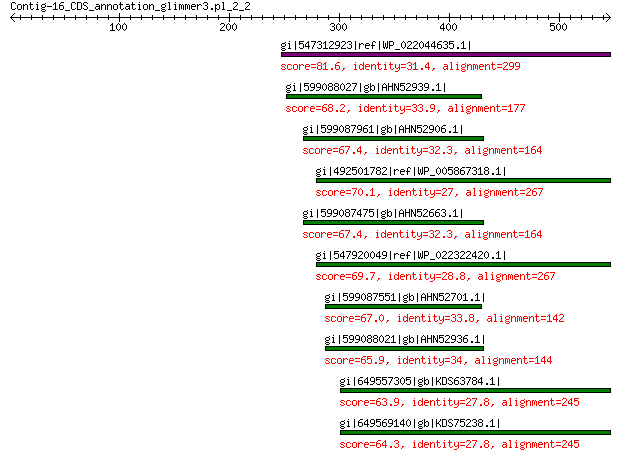
gi|547312923|ref|WP_022044635.1| putative uncharacterized protein 81.6 1e-13
gi|599088027|gb|AHN52939.1| major capsid protein 68.2 1e-09
gi|599087961|gb|AHN52906.1| major capsid protein 67.4 2e-09
gi|492501782|ref|WP_005867318.1| hypothetical protein 70.1 2e-09
gi|599087475|gb|AHN52663.1| major capsid protein 67.4 2e-09
gi|547920049|ref|WP_022322420.1| capsid protein VP1 69.7 2e-09
gi|599087551|gb|AHN52701.1| major capsid protein 67.0 3e-09
gi|599088021|gb|AHN52936.1| major capsid protein 65.9 6e-09
gi|649557305|gb|KDS63784.1| capsid family protein 63.9 3e-08
gi|649569140|gb|KDS75238.1| capsid family protein 64.3 7e-08
>gi|547312923|ref|WP_022044635.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
gi|524208404|emb|CCZ76639.1| putative uncharacterized protein [Alistipes finegoldii CAG:68]
Length=338
Score = 81.6 bits (200), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 94/336 (28%), Positives = 139/336 (41%), Gaps = 47/336 (14%)
Query 247 GLALKTYNSDLLQNWINTEWIEGEQGINEISA-----VDVSNG-QLTMDALNLAQKVYNM 300
GL Y+ DL N I +G EI +++S G + + L L K+ N
Sbjct 11 GLLSVPYSPDLFGNIIK----QGSSPAVEIEVMNALDLNISTGFSVAVPELRLRTKIQNW 66
Query 301 LNRIAVSGGTYRDWLETVFTGGNYMERCETPVFEGGMSQEIV---FQEVISNSASGEEP- 356
++R+ VSGG D T++ + P F G I + + + SASGE+
Sbjct 67 MDRLFVSGGRVGDVFRTLWGTKSSAIYVNKPDFLGVWQASINPSNVRAMANGSASGEDAN 126
Query 357 LGTLAGRGISTEKQKGGHVKIK--VTEPCYIIGIGSITPRIDYSQGNEFYAYHQTVDDIH 414
LG LA + GH I EP + I + P YSQG + D
Sbjct 127 LGQLAAC-VDRYCDFSGHSGIDYYAKEPGTFMLITMLVPEPAYSQGLHPDLASISFGDDF 185
Query 415 KPALDGIGYQDSVNWQRAFWDRQYNTTGQIQQPA------------------VGKTVAWI 456
P L+GIG+Q + + R +N TG Q+ + VG+ VAW
Sbjct 186 NPELNGIGFQLVPRHRFSMMPRGFNFTGLDQEASPWFGHTGTGVLVDPNMVSVGEEVAWS 245
Query 457 NYMTNINRTFGNFADNNSEAFMVMNRNYEYRSGTTF----GTTAIND---LTTYIDPVKF 509
T+ +R G+FA N + + V+ R + TT+ GT D TYI+P+ +
Sbjct 246 WLRTDYSRLHGDFAQNGNYQYWVLTRRF-----TTYFPDDGTGFYQDGEYTGTYINPLDW 300
Query 510 NYIFADTNLDAMNFWVQTKFDIKCRRLISAKQIPNL 545
Y+F D L A NF FD+ +SA +P L
Sbjct 301 QYVFVDQTLMAGNFAYYGTFDLNVTSSLSANYMPYL 336
>gi|599088027|gb|AHN52939.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=219
Score = 68.2 bits (165), Expect = 1e-09, Method: Compositional matrix adjust.
Identities = 60/180 (33%), Positives = 85/180 (47%), Gaps = 7/180 (4%)
Query 252 TYNSDLLQNWINTEWIEGEQGI--NEISAVDVSNG-QLTMDALNLAQKVYNMLNRIAVSG 308
T + +LLQ + G+ G+ N++ A D+S T++ L A ++ +L R A SG
Sbjct 40 TSSYELLQADQKYLFRPGDAGVQANQLYA-DLSQATAATINQLRQAFQIQKLLERDARSG 98
Query 309 GTYRDWLETVFTGGNYMERCETPVFEGGMSQEIVFQEVISNSASGEEPLGTLAGRGISTE 368
Y + ++ F G N+M+ P F GG S I V S SG P GTLA G +T
Sbjct 99 TRYAEIVKAHF-GVNFMDVTYRPEFLGGTSTPINVTSVPQTSESGTTPQGTLAAFGTATV 157
Query 369 KQKGGHVKIKVTEPCYIIGIGSITPRIDYSQGNEFYAYHQTVDDIHKPALDGIGYQDSVN 428
GG TE C ++GI S+ + Y QG T D + PAL IG Q +N
Sbjct 158 N--GGGFTKSFTEHCIVMGIASVRADLTYQQGLNRMFSRSTRYDFYFPALAHIGEQAVLN 215
>gi|599087961|gb|AHN52906.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=210
Score = 67.4 bits (163), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 53/164 (32%), Positives = 76/164 (46%), Gaps = 7/164 (4%)
Query 267 IEGEQGINEISAVDVSNGQLTMDALNLAQKVYNMLNRIAVSGGTYRDWLETVFTGGNYME 326
+G+Q ++S + T++ L A ++ +L R A SG Y + ++ F G N+M+
Sbjct 52 FDGQQLYTDLSTATAA----TINQLRQAFQIQKLLERDARSGTRYSEIVKAHF-GVNFMD 106
Query 327 RCETPVFEGGMSQEIVFQEVISNSASGEEPLGTLAGRGISTEKQKGGHVKIKVTEPCYII 386
P F GG S I V S SG P GTLA G +T GG TE C ++
Sbjct 107 VTYRPEFLGGTSTPINVTSVPQTSESGTTPQGTLAAFGTAT--INGGGFTKSFTEHCIVM 164
Query 387 GIGSITPRIDYSQGNEFYAYHQTVDDIHKPALDGIGYQDSVNWQ 430
GI S+ + Y QG T D + PAL IG Q +N +
Sbjct 165 GIASVRADLTYQQGLNRMFSRSTRYDFYFPALAHIGEQSVLNKE 208
>gi|492501782|ref|WP_005867318.1| hypothetical protein [Parabacteroides distasonis]
gi|409230408|gb|EKN23272.1| hypothetical protein HMPREF1059_03257 [Parabacteroides distasonis
CL09T03C24]
Length=538
Score = 70.1 bits (170), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 72/270 (27%), Positives = 110/270 (41%), Gaps = 23/270 (9%)
Query 279 VDVSNGQLTMDALNLAQKVYNMLNRIAVSGGTYRDWLETVFTGGNYMERCETPVFEGGMS 338
V+V ++++ L + + R A SG Y + + + F + R + P F GG
Sbjct 289 VNVDELGVSINDLRTSNALQRWFERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGR 348
Query 339 QEIVFQEVISNSAS-GEEPLGTLAGRGISTEKQKGGHVKIKVTEPCYIIGIGSITPRIDY 397
I EV+ SA+ P +AG GIS G K E YIIGI SI PR Y
Sbjct 349 TPISVSEVLQTSATDSTSPQANMAGHGISAGVNHG--FKRYFEEHGYIIGIMSIRPRTGY 406
Query 398 SQG--NEFYAYHQTVDDIHKPALDGIGYQDSVNWQRAFWDRQYNTTGQIQQPAVGKTVAW 455
QG +F + D + P +G Q+ N + + G G T +
Sbjct 407 QQGVPKDFRKFDNM--DFYFPEFAHLGEQEIKNEEVYLQQTPASNNGTF-----GYTPRY 459
Query 456 INYMTNINRTFGNFADNNSEAFMVMNRNYEYRSGTTFGTTAINDLTTYIDPVKFNYIFAD 515
Y ++N G+F N AF +NR + + + N TT+++ N +FA
Sbjct 460 AEYKYSMNEVHGDFRGN--MAFWHLNRIF---------SESPNLNTTFVECNPSNRVFAT 508
Query 516 TNLDAMNFWVQTKFDIKCRRLISAKQIPNL 545
+W+Q D+K RL+ P L
Sbjct 509 AETSDDKYWIQLYQDVKALRLMPKYGTPML 538
>gi|599087475|gb|AHN52663.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=210
Score = 67.4 bits (163), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 53/164 (32%), Positives = 76/164 (46%), Gaps = 7/164 (4%)
Query 267 IEGEQGINEISAVDVSNGQLTMDALNLAQKVYNMLNRIAVSGGTYRDWLETVFTGGNYME 326
+G+Q ++S + T++ L A ++ +L R A SG Y + ++ F G N+M+
Sbjct 52 FDGQQLYTDLSTATAA----TINQLRQAFQIQKLLERDARSGTRYSEIVKAHF-GVNFMD 106
Query 327 RCETPVFEGGMSQEIVFQEVISNSASGEEPLGTLAGRGISTEKQKGGHVKIKVTEPCYII 386
P F GG S I V S SG P GTLA G +T GG TE C ++
Sbjct 107 VTYRPEFLGGTSTPINVTSVPQTSESGTTPQGTLAAFGTAT--INGGGFTKSFTEHCILM 164
Query 387 GIGSITPRIDYSQGNEFYAYHQTVDDIHKPALDGIGYQDSVNWQ 430
GI S+ + Y QG T D + PAL IG Q +N +
Sbjct 165 GIASVRADLTYQQGLNRMFSRSTRYDFYFPALAHIGEQSVLNKE 208
>gi|547920049|ref|WP_022322420.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
gi|524592961|emb|CDD13573.1| capsid protein VP1 [Parabacteroides merdae CAG:48]
Length=553
Score = 69.7 bits (169), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 77/272 (28%), Positives = 112/272 (41%), Gaps = 27/272 (10%)
Query 279 VDVSNGQLTMDALNLAQKVYNMLNRIAVSGGTYRDWLETVFTGGNYMERCETPVFEGGMS 338
V+V + ++ L + + R A G Y + + + F + R + P F GG
Sbjct 304 VNVDEMGININDLRTSNALQRWFERNARGGSRYIEQILSHFGVRSSDARLQRPQFLGGGR 363
Query 339 QEIVFQEVISNSASGE-EPLGTLAGRGISTEKQKGGHVKIKVTEPCYIIGIGSITPRIDY 397
I EV+ S++ E P +AG GIS G K E YIIGI SITPR Y
Sbjct 364 MPISVSEVLQTSSTDETSPQANMAGHGISAGINNG--FKHYFEEHGYIIGIMSITPRSGY 421
Query 398 SQG--NEFYAYHQTVDDIHKPALDGIGYQDSVNWQRAFW--DRQYNTTGQIQQPAVGKTV 453
QG +F + D + P + Q+ N Q F D YN G T
Sbjct 422 QQGVPRDFTKFDNM--DFYFPEFAHLSEQEIKN-QELFVSEDAAYNNG------TFGYTP 472
Query 454 AWINYMTNINRTFGNFADNNSEAFMVMNRNYEYRSGTTFGTTAINDLTTYIDPVKFNYIF 513
+ Y + + G+F N S F +NR +E + N TT+++ N +F
Sbjct 473 RYAEYKYHPSEAHGDFRGNLS--FWHLNRIFEDKP---------NLNTTFVECKPSNRVF 521
Query 514 ADTNLDAMNFWVQTKFDIKCRRLISAKQIPNL 545
A + + FWVQ D+K RL+ P L
Sbjct 522 ATSETEDDKFWVQMYQDVKALRLMPKYGTPML 553
>gi|599087551|gb|AHN52701.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=220
Score = 67.0 bits (162), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 48/142 (34%), Positives = 70/142 (49%), Gaps = 2/142 (1%)
Query 287 TMDALNLAQKVYNMLNRIAVSGGTYRDWLETVFTGGNYMERCETPVFEGGMSQEIVFQEV 346
T++ L A ++ +L R A G Y + ++ F + R + P + GG + I+ +V
Sbjct 77 TINQLRQAFQIQKLLERDARGGTRYTEIIQAHFGVTSPDARLQRPEYLGGGTTPIIISQV 136
Query 347 ISNSASGEEPLGTLAGRGISTEKQKGGHVKIKVTEPCYIIGIGSITPRIDYSQGNEFYAY 406
S S P GTLA G +T + K G K TE C IIG+ S+ + Y QG E
Sbjct 137 PQTSESDGTPQGTLAAYGTATMR-KAGFTK-SFTEHCVIIGLASVRADLTYQQGLERMWS 194
Query 407 HQTVDDIHKPALDGIGYQDSVN 428
QT D++ PAL IG Q +N
Sbjct 195 RQTRYDVYWPALAMIGEQAVLN 216
>gi|599088021|gb|AHN52936.1| major capsid protein, partial [uncultured Gokushovirinae]
Length=220
Score = 65.9 bits (159), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 49/144 (34%), Positives = 68/144 (47%), Gaps = 3/144 (2%)
Query 287 TMDALNLAQKVYNMLNRIAVSGGTYRDWLETVFTGGNYMERCETPVFEGGMSQEIVFQEV 346
T++ L A ++ +L R A SG Y + ++ F G N+M+ P F GG S + V
Sbjct 78 TINQLRQAFQIQKLLERDARSGTRYSEIVKAHF-GVNFMDVTYRPEFLGGSSTPVNVTSV 136
Query 347 ISNSASGEEPLGTLAGRGISTEKQKGGHVKIKVTEPCYIIGIGSITPRIDYSQGNEFYAY 406
S SG P GTLA G +T GG TE C ++GI S+ + Y QG
Sbjct 137 PQTSESGTTPQGTLAAFGTAT--INGGGFTKSFTEHCIVMGIASVRADLTYQQGLNRMFS 194
Query 407 HQTVDDIHKPALDGIGYQDSVNWQ 430
T D + PAL IG Q +N +
Sbjct 195 RSTRYDFYFPALAHIGEQSVLNKE 218
>gi|649557305|gb|KDS63784.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 4]
gi|649559156|gb|KDS65543.1| capsid family protein [Parabacteroides distasonis str. 3999B
T(B) 6]
Length=245
Score = 63.9 bits (154), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 68/248 (27%), Positives = 99/248 (40%), Gaps = 23/248 (9%)
Query 301 LNRIAVSGGTYRDWLETVFTGGNYMERCETPVFEGGMSQEIVFQEVISNSASGE-EPLGT 359
R A SG Y + + + F + R + P F GG I EV+ S++ P
Sbjct 18 FERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQAN 77
Query 360 LAGRGISTEKQKGGHVKIKVTEPCYIIGIGSITPRIDYSQG--NEFYAYHQTVDDIHKPA 417
+AG GIS G E YI+GI SI PR Y QG +F + D + P
Sbjct 78 MAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNM--DFYFPE 133
Query 418 LDGIGYQDSVNWQRAFWDRQYNTTGQIQQPAVGKTVAWINYMTNINRTFGNFADNNSEAF 477
+G Q+ N + N + + G T + Y + N G+F N AF
Sbjct 134 FAHLGEQEIKNEELYL-----NESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGN--MAF 186
Query 478 MVMNRNYEYRSGTTFGTTAINDLTTYIDPVKFNYIFADTNLDAMNFWVQTKFDIKCRRLI 537
+NR ++ + N TT+++ N +FA +WVQ DIK RL+
Sbjct 187 WHLNRIFKEKP---------NLNTTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLM 237
Query 538 SAKQIPNL 545
P L
Sbjct 238 PKYGTPML 245
>gi|649569140|gb|KDS75238.1| capsid family protein, partial [Parabacteroides distasonis str.
3999B T(B) 6]
Length=390
Score = 64.3 bits (155), Expect = 7e-08, Method: Compositional matrix adjust.
Identities = 68/248 (27%), Positives = 99/248 (40%), Gaps = 23/248 (9%)
Query 301 LNRIAVSGGTYRDWLETVFTGGNYMERCETPVFEGGMSQEIVFQEVISNSAS-GEEPLGT 359
R A SG Y + + + F + R + P F GG I EV+ S++ P
Sbjct 163 FERNARSGSRYIEQILSHFGVRSSDARLQRPQFLGGGRTPISVSEVLQTSSTDSTSPQAN 222
Query 360 LAGRGISTEKQKGGHVKIKVTEPCYIIGIGSITPRIDYSQG--NEFYAYHQTVDDIHKPA 417
+AG GIS G E YI+GI SI PR Y QG +F + D + P
Sbjct 223 MAGHGISAGVNHG--FTRYFEEHGYIMGIMSIRPRTGYQQGVPKDFRKFDNM--DFYFPE 278
Query 418 LDGIGYQDSVNWQRAFWDRQYNTTGQIQQPAVGKTVAWINYMTNINRTFGNFADNNSEAF 477
+G Q+ N + N + + G T + Y + N G+F N AF
Sbjct 279 FAHLGEQEIKNEELYL-----NESDAANEGTFGYTPRYAEYKYSQNEVHGDFRGN--MAF 331
Query 478 MVMNRNYEYRSGTTFGTTAINDLTTYIDPVKFNYIFADTNLDAMNFWVQTKFDIKCRRLI 537
+NR ++ + N TT+++ N +FA +WVQ DIK RL+
Sbjct 332 WHLNRIFKEKP---------NLNTTFVECNPSNRVFATAETSDDKYWVQIYQDIKALRLM 382
Query 538 SAKQIPNL 545
P L
Sbjct 383 PKYGTPML 390
Lambda K H a alpha
0.316 0.133 0.395 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3945317559120