bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_5
Length=57
Score E
Sequences producing significant alignments: (Bits) Value
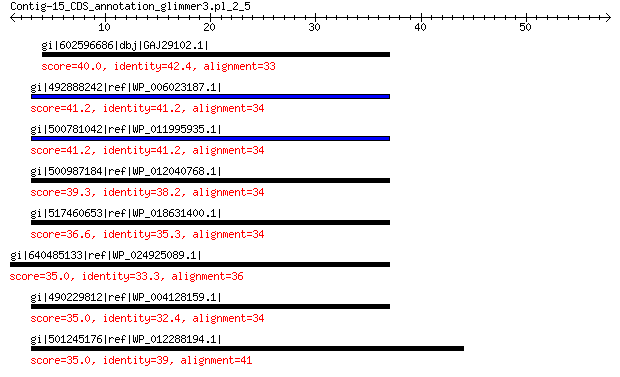
gi|602596686|dbj|GAJ29102.1| resolvase 40.0 0.017
gi|492888242|ref|WP_006023187.1| hypothetical protein 41.2 0.024
gi|500781042|ref|WP_011995935.1| serine recombinase 41.2 0.024
gi|500987184|ref|WP_012040768.1| serine recombinase 39.3 0.093
gi|517460653|ref|WP_018631400.1| serine recombinase 36.6 0.67
gi|640485133|ref|WP_024925089.1| MULTISPECIES: serine recombinase 35.0 2.3
gi|490229812|ref|WP_004128159.1| resolvase domain-containing pro... 35.0 2.4
gi|501245176|ref|WP_012288194.1| serine recombinase 35.0 2.4
>gi|602596686|dbj|GAJ29102.1| resolvase [Acidomonas methanolica NBRC 104435]
Length=180
Score = 40.0 bits (92), Expect = 0.017, Method: Composition-based stats.
Identities = 14/33 (42%), Positives = 25/33 (76%), Gaps = 0/33 (0%)
Query 4 YYLCSIQSKLNPSQCETVLVPVDEVSDFVSSNL 36
YY CS++++ P+ CE + VP+D++ D V+S+L
Sbjct 27 YYACSMKARQGPTACEGMAVPMDKLDDLVASHL 59
>gi|492888242|ref|WP_006023187.1| hypothetical protein [Afipia broomeae]
gi|410886764|gb|EKS34576.1| hypothetical protein HMPREF9695_04486 [Afipia broomeae ATCC 49717]
Length=540
Score = 41.2 bits (95), Expect = 0.024, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 26/34 (76%), Gaps = 0/34 (0%)
Query 3 KYYLCSIQSKLNPSQCETVLVPVDEVSDFVSSNL 36
+YY CS++++ P+ CE + VP+D++ D V+S+L
Sbjct 329 RYYACSMKARQGPTACEGMAVPMDKLDDLVASHL 362
>gi|500781042|ref|WP_011995935.1| serine recombinase [Xanthobacter autotrophicus]
gi|154244234|ref|YP_001415192.1| resolvase domain-containing protein [Xanthobacter autotrophicus
Py2]
gi|154158319|gb|ABS65535.1| Resolvase domain [Xanthobacter autotrophicus Py2]
Length=551
Score = 41.2 bits (95), Expect = 0.024, Method: Composition-based stats.
Identities = 14/34 (41%), Positives = 26/34 (76%), Gaps = 0/34 (0%)
Query 3 KYYLCSIQSKLNPSQCETVLVPVDEVSDFVSSNL 36
+YY CS++++ P+ CE + VP+D++ D V+S+L
Sbjct 329 RYYACSMKARQGPTACEGMAVPMDKLDDLVASHL 362
>gi|500987184|ref|WP_012040768.1| serine recombinase [Bradyrhizobium sp. BTAi1]
gi|148252037|ref|YP_001236622.1| resolvase [Bradyrhizobium sp. BTAi1]
gi|146404210|gb|ABQ32716.1| putative Resolvase [Bradyrhizobium sp. BTAi1]
Length=540
Score = 39.3 bits (90), Expect = 0.093, Method: Composition-based stats.
Identities = 13/34 (38%), Positives = 26/34 (76%), Gaps = 0/34 (0%)
Query 3 KYYLCSIQSKLNPSQCETVLVPVDEVSDFVSSNL 36
+YY CS++++ P+ CE + VP+D++ + V+S+L
Sbjct 329 RYYACSMKARQGPTACEGLAVPMDKLDELVASHL 362
>gi|517460653|ref|WP_018631400.1| serine recombinase [Meganema perideroedes]
Length=549
Score = 36.6 bits (83), Expect = 0.67, Method: Composition-based stats.
Identities = 12/34 (35%), Positives = 25/34 (74%), Gaps = 0/34 (0%)
Query 3 KYYLCSIQSKLNPSQCETVLVPVDEVSDFVSSNL 36
+YY CS++++ P+ CE + VP++++ D V ++L
Sbjct 332 RYYACSMKARQGPTACEGMAVPMEKLDDLVVNHL 365
>gi|640485133|ref|WP_024925089.1| MULTISPECIES: serine recombinase [Mesorhizobium]
Length=552
Score = 35.0 bits (79), Expect = 2.3, Method: Composition-based stats.
Identities = 12/36 (33%), Positives = 24/36 (67%), Gaps = 0/36 (0%)
Query 1 MEKYYLCSIQSKLNPSQCETVLVPVDEVSDFVSSNL 36
M +YY CS +++ + CE +P+D++ D ++S+L
Sbjct 336 MYRYYTCSTKARQGKTGCEGRSIPMDKLDDLIASHL 371
>gi|490229812|ref|WP_004128159.1| resolvase domain-containing protein [Rhizobium freirei]
gi|478272602|gb|ENN84553.1| resolvase domain-containing protein [Rhizobium freirei PRF 81]
Length=597
Score = 35.0 bits (79), Expect = 2.4, Method: Composition-based stats.
Identities = 11/34 (32%), Positives = 24/34 (71%), Gaps = 0/34 (0%)
Query 3 KYYLCSIQSKLNPSQCETVLVPVDEVSDFVSSNL 36
+YY CS++++ P+ C + VP++++ D V+ +L
Sbjct 334 RYYACSMKARQGPTACAGMAVPMEKLDDLVAGHL 367
>gi|501245176|ref|WP_012288194.1| serine recombinase [Caulobacter sp. K31]
gi|167648141|ref|YP_001685804.1| resolvase domain-containing protein [Caulobacter sp. K31]
gi|167350571|gb|ABZ73306.1| Resolvase domain [Caulobacter sp. K31]
Length=579
Score = 35.0 bits (79), Expect = 2.4, Method: Composition-based stats.
Identities = 16/48 (33%), Positives = 30/48 (63%), Gaps = 7/48 (15%)
Query 3 KYYLCSIQSKLNPSQCETVLVPVDEVSDFVSSN-----LRPD--CILI 43
+YY CS ++++ S C + VP+D++ D V ++ L+PD C+L+
Sbjct 339 RYYTCSTKARMGKSGCTGITVPMDKLDDAVVNHLERRLLQPDRLCMLM 386
Lambda K H a alpha
0.320 0.132 0.394 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 434510049843