bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: All non-redundant GenBank CDS translations+PDB+SwissProt+PIR+PRF
excluding environmental samples from WGS projects
49,011,213 sequences; 17,563,301,199 total letters
Query= Contig-12_CDS_annotation_glimmer3.pl_2_5
Length=623
Score E
Sequences producing significant alignments: (Bits) Value
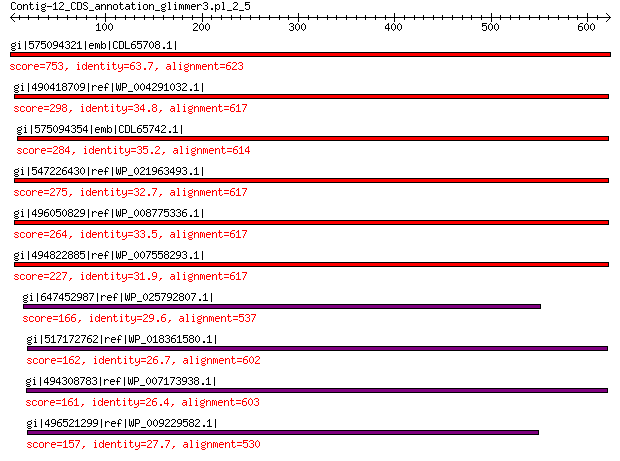
gi|575094321|emb|CDL65708.1| unnamed protein product 753 0.0
gi|490418709|ref|WP_004291032.1| hypothetical protein 298 1e-88
gi|575094354|emb|CDL65742.1| unnamed protein product 284 4e-83
gi|547226430|ref|WP_021963493.1| putative uncharacterized protein 275 8e-80
gi|496050829|ref|WP_008775336.1| hypothetical protein 264 6e-76
gi|494822885|ref|WP_007558293.1| hypothetical protein 227 6e-62
gi|647452987|ref|WP_025792807.1| hypothetical protein 166 8e-41
gi|517172762|ref|WP_018361580.1| hypothetical protein 162 1e-39
gi|494308783|ref|WP_007173938.1| hypothetical protein 161 2e-39
gi|496521299|ref|WP_009229582.1| capsid protein 157 7e-38
>gi|575094321|emb|CDL65708.1| unnamed protein product [uncultured bacterium]
Length=642
Score = 753 bits (1945), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 397/655 (61%), Positives = 469/655 (72%), Gaps = 45/655 (7%)
Query 1 MANRSNIMGLHGLKNKTSRNSFDLSHRNLFTAKVGELLPCFVQEVNPGDSIKLDSSYFTR 60
MANRSNIMGLHGLKNK SRNSFDLSHRN+FTAKVGELLPCFVQE+NPGDS+K+ SSYFTR
Sbjct 1 MANRSNIMGLHGLKNKPSRNSFDLSHRNMFTAKVGELLPCFVQELNPGDSVKVSSSYFTR 60
Query 61 TAPLQTAAFTRLRENVQYFFVPYQCLWKYFEGQVKNMTKNANGGDISQIATSPFANAKVS 120
TAPLQ+ AFTRLRENVQYFFVPY LWKYF+ QV NMTKNANGGDIS+IA+S N KV+
Sbjct 61 TAPLQSNAFTRLRENVQYFFVPYSALWKYFDSQVLNMTKNANGGDISRIASSLVGNQKVT 120
Query 121 TEMPFISYTALHAYLNKLLNY----VDSSANPTELSNPFLYNNGCWRHAESAKLLQLLGY 176
T+MP ++Y LHAYL K +N D S P +N GC+RHAESAKLLQLLGY
Sbjct 121 TQMPCVNYKTLHAYLLKFINRSTVGSDGSVGPE-------FNRGCYRHAESAKLLQLLGY 173
Query 177 GNFVQQFKNFS--------ASKPYSLLHVENAPALSVFRLLAYQKICNDFYTYRQWQPYN 228
GNF +QF NF + + + + N+P LS+FRLLAY KICND Y YRQWQPYN
Sbjct 174 GNFPEQFANFKVNNDKHNQSGQNFKDVTYNNSPYLSIFRLLAYHKICNDHYLYRQWQPYN 233
Query 229 ASLCNIDYITPDssssmdlsskfssisVSDLG--KSNMLDMRFSNLPLDYFNGVLPTPQF 286
ASLCN+DY+TP+SSS + + SI + K N+LDMRFSNLPLDYF GVLPT QF
Sbjct 234 ASLCNVDYLTPNSSSLLSIDDALLSIPDDSIKAEKLNLLDMRFSNLPLDYFTGVLPTSQF 293
Query 287 GSESVVSL-------SQNADVYTGFDKSQWQTLDG--safpsgsvsssnsdrsltANGKS 337
GSESVV+L S + T D +W+T G + S++ + + +NG
Sbjct 294 GSESVVNLNLGNASGSAVLNGTTSKDSGRWRTTTGEWEMEQRVASSANGNLKLDNSNGTF 353
Query 338 IEHVHILPSG-----SITSSLSIAALRQATALQKYKEIQLANDPDFESQIEAHFGIKPKH 392
I H H S++ +LSI ALR A A QKYKEIQLAND DF+SQ+EAHFGIKP
Sbjct 354 ISHDHTFSGNVAINTSLSGNLSIIALRNALAAQKYKEIQLANDVDFQSQVEAHFGIKPDE 413
Query 393 DMHKSRFIGGSSSMIDINPVVNQNLGAGQNQDNQAVTKAAPTGQGGASFKFTADTFGVVI 452
S FIGGSSSMI+IN +NQNL DN+A AAP G G AS KFTA T+GVVI
Sbjct 414 KNENSLFIGGSSSMININEQINQNLSG----DNKATYGAAPQGNGSASIKFTAKTYGVVI 469
Query 453 GIYRCTPVLDYSHVGIDRTLLKTDASDFVIPELDSIGMQQTFQCELFAPT---SQMTA-S 508
GIYRCTPVLD++H+GIDRTL KTDASDFVIPE+DSIGMQQTF+CE+ AP + A
Sbjct 470 GIYRCTPVLDFAHLGIDRTLFKTDASDFVIPEMDSIGMQQTFRCEVAAPAPYNDEFKAFR 529
Query 509 APDKRKYDMSRTFGYAPRYSEYKVSFDRYNGAFCDTLKSWVTGFNTHIFDSDRWNDRSYF 568
D DMS T+GYAPRYSE+K S+DRYNGAFC +LKSWVTG N ++ WN ++
Sbjct 530 VGDGSSPDMSETYGYAPRYSEFKTSYDRYNGAFCHSLKSWVTGINFDAIQNNVWN--TWA 587
Query 569 SISVPQLFVCRPDIVKDIFALQTYHDSNDDNLYVGMVNMCYATRNLSRYGLPYSN 623
I+ P +F CRPDIVK++F + + ++S+DD LYVGMVNMCYATRNLSRYGLPYSN
Sbjct 588 GINAPNMFACRPDIVKNLFLVSSTNNSDDDQLYVGMVNMCYATRNLSRYGLPYSN 642
>gi|490418709|ref|WP_004291032.1| hypothetical protein [Bacteroides eggerthii]
gi|217986636|gb|EEC52970.1| putative capsid protein (F protein) [Bacteroides eggerthii DSM
20697]
Length=578
Score = 298 bits (762), Expect = 1e-88, Method: Compositional matrix adjust.
Identities = 215/631 (34%), Positives = 302/631 (48%), Gaps = 68/631 (11%)
Query 5 SNIMGLHGLKNKTSRNSFDLSHRNLFTAKVGELLPCFVQEVNPGDSIKLDSSYFTRTAPL 64
+NIM L ++NK SRN FDLS + FTAK GELLP V+EV PGD+ K++ FTRT P+
Sbjct 2 ANIMSLKSIRNKPSRNGFDLSFKKNFTAKAGELLPVMVKEVLPGDTFKINLKAFTRTQPV 61
Query 65 QTAAFTRLRENVQYFFVPYQCLWKYFEGQVKNMTKNANGGDISQIATSPFANAKVSTEMP 124
TAAF R+RE +FFVPY LW + M N ++ P N +S EMP
Sbjct 62 NTAAFARIREYYDFFFVPYDLLWNKANTVLTQMYDNPQHA----VSIDPTRNFVLSGEMP 117
Query 125 FISYTALHAYLNKLLNYVDSSANPTELSNPFLYNNGCWRHAESAKLLQLLGYGNFVQQFK 184
+++ A+ +Y+N L +SA SN F YN R S KLL+ LGYGN+
Sbjct 118 YMTSEAIASYINALST---ASALADYKSNYFGYN----RSKSSVKLLEYLGYGNYESFLT 170
Query 185 NFSASKPY--SLLHVENAPALSVFRLLAYQKICNDFYTYRQWQPYNASLCNIDYITPDss 242
+ + P +L H ++F LLAYQKI +DFY QW+ + S N+DY+ S
Sbjct 171 DDWNTAPLMANLNH-------NIFGLLAYQKIYSDFYRDSQWERVSPSTFNVDYLDGSSM 223
Query 243 ssmdlsskfssisVSDLGKSNMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYT 302
+ + S N D+R+ N D F+GVLP Q+G +V S++ DV
Sbjct 224 NLDNAYSTEFYQ------NYNFFDLRYCNWQKDLFHGVLPHQQYGETAVASIT--PDVTG 275
Query 303 GFDKSQWQTLDGsafpsgsvsssnsdrsltANGKSIEHVHILPSGSITSSLSIAALRQAT 362
S + T+ TA+G + ++ LP+ LSI LRQA
Sbjct 276 KLTLSNFSTV--------------GTSPTTASGTATKN---LPAFDTVGDLSILVLRQAE 318
Query 363 ALQKYKEIQLANDPDFESQIEAHFGIKPKHDMHK-SRFIGGSSSMIDINPVVNQNLGAGQ 421
LQK+KEI + + D++ Q+E H+G+ + ++GG SS IDIN V+N N+
Sbjct 319 FLQKWKEITQSGNKDYKDQLEKHWGVSVGDGFSELCTYLGGVSSSIDINEVINTNITGSA 378
Query 422 NQDNQAVTKAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFV 481
D K G +F + +G+++ IY C P+LDY+ +D LK +++D+
Sbjct 379 AAD--IAGKGVGVANGEINFN-SNGRYGLIMCIYHCLPLLDYTTDMLDPAFLKVNSTDYA 435
Query 482 IPELDSIGMQQTFQCELFAPTSQMTASAPDKRKYDMSRTFGYAPRYSEYKVSFDRYNGAF 541
IPE D +GMQ +L P ++ GY PRY +YK S D+ G F
Sbjct 436 IPEFDRVGMQSMPLVQLMNPLRSFANAS--------GLVLGYVPRYIDYKTSVDQSVGGF 487
Query 542 CDTLKSWVTGF-NTHIFDSDRW-NDRSYFSISVP---------QLFVCRPDIVKDIFALQ 590
TL SWV + N + ND S P F PD + IFA+Q
Sbjct 488 KRTLNSWVISYGNISVLKQVTLPNDAPPIEPSEPVPSVAPMNFTFFKVNPDCLDPIFAVQ 547
Query 591 TYHDSNDDNLYVGMVNMCYATRNLSRYGLPY 621
D+N D A RNL GLPY
Sbjct 548 AGDDTNTDQFLCSSFFDIKAVRNLDTDGLPY 578
>gi|575094354|emb|CDL65742.1| unnamed protein product [uncultured bacterium]
Length=615
Score = 284 bits (727), Expect = 4e-83, Method: Compositional matrix adjust.
Identities = 216/662 (33%), Positives = 310/662 (47%), Gaps = 95/662 (14%)
Query 8 MGLHGLKNKTSRNSFDLSHRNLFTAKVGELLPCFVQEVNPGDSIKLDSSYFTRTAPLQTA 67
M + +KN+ SRN FDLS + FTAK GELLP + V PGDS ++ FTRT PL T+
Sbjct 1 MSMADIKNRPSRNGFDLSFKKNFTAKAGELLPVMTKVVLPGDSFNINLRSFTRTQPLNTS 60
Query 68 AFTRLRENVQYFFVPYQCLWKYFEGQVKNMTKNANGGDISQIATSPFA--NAKVSTEMPF 125
AF R+RE ++FVP++ +W F+ + M N Q A+ P N +S MP+
Sbjct 61 AFARMREYYDFYFVPFEQMWNKFDSCITQMNANV------QHASGPTLDDNTPLSGRMPY 114
Query 126 ISYTALHAYLNKLLNYVDSSANPTELSNPFLYNNGCWRHAESAKLLQLLGYGNF--VQQF 183
+ + YLN NPF +N R + KLLQ LGYG++
Sbjct 115 FTSEQIADYLNDQAT--------AARKNPFGFN----RSTLTCKLLQYLGYGDYNSFDSE 162
Query 184 KNFSASKPYSLLHVENAPALSVFRLLAYQKICNDFYTYRQWQPYNASLCNIDYITPDsss 243
N ++KP L ++E LS F LLAYQKI +DFY Y QW+ N S N+DYI
Sbjct 163 TNTWSAKPL-LYNLE----LSPFPLLAYQKIYSDFYRYTQWEKTNPSTFNLDYI-----K 212
Query 244 smdlsskfssisVSDLGKSNMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYTG 303
+ SD +N D+R+ N D F+GVLP Q+GS SVV ++ +V +
Sbjct 213 GTSDLQMDLTGLPSD--DNNFFDIRYCNYQKDMFHGVLPVAQYGSASVVPINGQLNVISN 270
Query 304 FDK---------------SQWQTLDGsafpsgsvsssnsdrsltANGKSIE-HVHILPSG 347
D + + T+ G+ + GKS + + PS
Sbjct 271 GDSGPIFKTSTPDPGTPGTSYVTVGGNIGVDNRSFGVSGSTLNV--GKSADPSGYGFPSN 328
Query 348 SITSSL-----------------SIAALRQATALQKYKEIQLANDPDFESQIEAHFGIKP 390
+ T SL I ALRQA LQK+KE+ ++ + D++SQIE H+GIK
Sbjct 329 ASTRSLLWENPNLIIENNQGFYVPILALRQAEFLQKWKEVSVSGEEDYKSQIEKHWGIKV 388
Query 391 KHDM-HKSRFIGGSSSMIDINPVVNQNLGAGQNQDNQAVTKAAPTGQGGASFKFTAD-TF 448
+ H++R++GG ++ +DIN V+N N+ DN A T G S +F + +
Sbjct 389 SDFLSHQARYLGGCATSLDINEVINNNITG----DNAADIAGKGTFTGNGSIRFESKGEY 444
Query 449 GVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFVIPELDSIGMQQTFQCELFAPTSQMTAS 508
G+++ IY P++DY G+D + DA+ F IPELD IGM+ P +
Sbjct 445 GIIMCIYHVLPIVDYVGSGVDHSCTLVDATSFPIPELDQIGMESVPLVRAMNPVKESDTP 504
Query 509 APDKRKYDMSRTF-GYAPRYSEYKVSFDRYNGAFCDTLKSW--------VTGFNTHIFDS 559
+ D TF GYAPRY ++K S DR G F D+L++W +T N+ F S
Sbjct 505 SAD--------TFLGYAPRYIDWKTSVDRSVGDFADSLRTWCLPVGDKELTSANSLNFPS 556
Query 560 DRWNDRSYFSISVPQLFVCRPDIVKDIFALQTYHDSNDDNLYVGMVNMCYATRNLSRYGL 619
+ + + F P IV +FA+ D RNL GL
Sbjct 557 NPNVEPDSIAAG---FFKVNPSIVDPLFAVVADSTVKTDEFLCSSFFDVKVVRNLDVNGL 613
Query 620 PY 621
PY
Sbjct 614 PY 615
>gi|547226430|ref|WP_021963493.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
gi|524103382|emb|CCY83994.1| putative uncharacterized protein [Prevotella sp. CAG:1185]
Length=573
Score = 275 bits (702), Expect = 8e-80, Method: Compositional matrix adjust.
Identities = 202/623 (32%), Positives = 292/623 (47%), Gaps = 57/623 (9%)
Query 5 SNIMGLHGLKNKTSRNSFDLSHRNLFTAKVGELLPCFVQEVNPGDSIKLDSSYFTRTAPL 64
S++M L LKN RN FDLS +N FTAKVGELLP +EV PGD + FTRT P+
Sbjct 2 SSVMSLTALKNSVKRNGFDLSFKNAFTAKVGELLPIMCKEVYPGDKFNIRGQAFTRTQPV 61
Query 65 QTAAFTRLRENVQYFFVPYQCLWKYFEGQVKNMTKNANGGDISQIATSPFANAKVSTEMP 124
+AA++RLRE ++FVPY+ LW NM + D+ ++ +S P
Sbjct 62 NSAAYSRLREYYDFYFVPYRLLWNMAPTFFTNMPDPHHAADL-------VSSVNLSQRHP 114
Query 125 FISYTALHAYLNKLLNYVDSSANPTELSNPFLYNNGCWRHAESAKLLQLLGYGNFVQQFK 184
+ ++ + YL L + S A N F G R S KLL L YG F + ++
Sbjct 115 WFTFFDIMEYLGNLNSL--SGAYEKYQKNFF----GFSRVELSVKLLNYLNYG-FGKDYE 167
Query 185 NFSASKPYSLLHVENAPALSVFRLLAYQKICNDFYTYRQWQPYNASLCNIDYITPDssss 244
+ + LS F LLAYQKIC D++ QWQ N+DY+
Sbjct 168 SVKVPSD------SDDIVLSPFPLLAYQKICEDYFRDDQWQSAAPYRYNLDYL----YGK 217
Query 245 mdlsskfssisVSDLGKS-NMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYTG 303
S +D K+ M D+ + N DYF G+LP Q+G SV S ++
Sbjct 218 SSGFHIPMSSFTNDAFKNPTMFDLNYCNFQKDYFTGMLPRAQYGDVSVAS-----PIFGD 272
Query 304 FDKSQWQTLDGsafpsgsvsssnsdrsltANGKSIEHVHILPSGSITSSLSIAALRQATA 363
D +L + S AN + + + + T+ LS+ ALRQA
Sbjct 273 LDIGDSSSL-----------TFASAPQQGANTIQSGVLVVNNNSNTTAGLSVLALRQAEC 321
Query 364 LQKYKEIQLANDPDFESQIEAHFGIKPKHDMH-KSRFIGGSSSMIDINPVVNQNLGAGQN 422
LQK++EI + D+++Q++ HF + P + +++GG +S +DI+ VVN NL
Sbjct 322 LQKWREIAQSGKMDYQTQMQKHFNVSPSATLSGHCKYLGGWTSNLDISEVVNTNLTG--- 378
Query 423 QDNQAVTKAAPTGQ-GGASFKFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFV 481
DNQA + TG G F + G+++ IY C P+LD+S I R KT +D+
Sbjct 379 -DNQADIQGKGTGTLNGNKVDFESSEHGIIMCIYHCLPLLDWSINRIARQNFKTTFTDYA 437
Query 482 IPELDSIGMQQTFQCELFAPTSQMTASAPDKRKYDMSRTFGYAPRYSEYKVSFDRYNGAF 541
IPE DS+GMQQ + E+ + + S GY PRY++ K S D +G+F
Sbjct 438 IPEFDSVGMQQLYPSEMIFGLEDLPSDPS-------SINMGYVPRYADLKTSIDEIHGSF 490
Query 542 CDTLKSWVTGFNTHIFDSDR--WNDRSYFSISVP-QLFVCRPDIVKDIFALQTYHDSNDD 598
DTL SWV+ + R D + I++ F P IV +IF ++ N D
Sbjct 491 IDTLVSWVSPLTDSYISAYRQACKDAGFSDITMTYNFFKVNPHIVDNIFGVKADSTINTD 550
Query 599 NLYVGMVNMCYATRNLSRYGLPY 621
L + A RN GLPY
Sbjct 551 QLLINSYFDIKAVRNFDYNGLPY 573
>gi|496050829|ref|WP_008775336.1| hypothetical protein [Bacteroides sp. 2_2_4]
gi|229448893|gb|EEO54684.1| putative capsid protein (F protein) [Bacteroides sp. 2_2_4]
Length=580
Score = 264 bits (675), Expect = 6e-76, Method: Compositional matrix adjust.
Identities = 207/625 (33%), Positives = 306/625 (49%), Gaps = 54/625 (9%)
Query 5 SNIMGLHGLKNKTSRNSFDLSHRNLFTAKVGELLPCFVQEVNPGDSIKLDSSYFTRTAPL 64
+NIM L L+NKTSRN FDLS + FTAK GELLP EV PGD +D FTRT PL
Sbjct 2 ANIMSLKSLRNKTSRNGFDLSSKRNFTAKPGELLPVKCWEVLPGDKWSIDLKSFTRTQPL 61
Query 65 QTAAFTRLRENVQYFFVPYQCLWKYFEGQVKNMTKNANGGDISQIATS--PFANAKVSTE 122
TAAF R+RE ++FVPY LW + M N Q ATS P AN ++
Sbjct 62 NTAAFARMREYYDFYFVPYNLLWNKANTVLTQMYDNP------QHATSYIPSANQALAGV 115
Query 123 MPFISYTALHAYLNKLLNYVDSSANPTELSNPFLYNNGCWRHAESAKLLQLLGYGNFVQQ 182
MP ++ + YLN L D + + N F Y+ R +AKLL+ LGYGNF
Sbjct 116 MPNVTCKGIADYLN--LVAPDVTTTNSYEKNYFGYS----RSLGTAKLLEYLGYGNFYTY 169
Query 183 FKNFSASKPYSLLHVENAPALSVFRLLAYQKICNDFYTYRQWQPYNASLCNIDYITPDss 242
S + ++ + + L+++ +LAYQKI D QW+ + S N+DY++
Sbjct 170 AT--SKNNTWTKSPLSSNLQLNIYGVLAYQKIYADHIRDSQWEKVSPSCFNVDYLSGTVD 227
Query 243 ssmdlsskfssisVSDLGKSNMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYT 302
S+M + S + + NM D+R+ N D F+GVLP Q+G + V+++ ++V +
Sbjct 228 SAMTIDSMITGQGFAPF--YNMFDLRYCNWQKDLFHGVLPRQQYGDTAAVNVNL-SNVLS 284
Query 303 GFDKSQWQTLDGsafpsgsvsssnsdrsltANGKSIEHVHILPSGSITSSLSIAALRQAT 362
+ QT DG ++ G +++ V+ SG+ T + ALRQA
Sbjct 285 A--QYMVQTPDG---------DPVGGSPFSSTGVNLQTVN--GSGTFT----VLALRQAE 327
Query 363 ALQKYKEIQLANDPDFESQIEAHFGIKPKHDMHK-SRFIGGSSSMIDINPVVNQNLGAGQ 421
LQK+KEI + + D++ QIE H+ + + S ++GG+++ +DIN VVN N+
Sbjct 328 FLQKWKEITQSGNKDYKDQIEKHWNVSVGEAYSEMSLYLGGTTASLDINEVVNNNITGSN 387
Query 422 NQDNQAVTKAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFV 481
D K G G SF + +G+++ IY P+LDY+ ++ K +++DF
Sbjct 388 AAD--IAGKGVVVGNGRISFD-AGERYGLIMCIYHSLPLLDYTTDLVNPAFTKINSTDFA 444
Query 482 IPELDSIGMQQTFQCELFAPTSQMTASAPDKRKYDM-SRTFGYAPRYSEYKVSFDRYNGA 540
IPE D +GM+ L P + Y++ S GYAPRY YK D GA
Sbjct 445 IPEFDRVGMESVPLVSLMNPL---------QSSYNVGSSILGYAPRYISYKTDVDSSVGA 495
Query 541 FCDTLKSWVTGF-NTHIFDSDRWND---RSYFSISVPQLFVCRPDIVKDIFALQTYHDSN 596
F TLKSWV + N + + + D S ++ F P+ V +FA+ + +
Sbjct 496 FKTTLKSWVMSYDNQSVINQLNYQDDPNNSPGTLVNYTNFKVNPNCVDPLFAVAASNSID 555
Query 597 DDNLYVGMVNMCYATRNLSRYGLPY 621
D RNL GLPY
Sbjct 556 TDQFLCSSFFDVKVVRNLDTDGLPY 580
>gi|494822885|ref|WP_007558293.1| hypothetical protein [Bacteroides plebeius]
gi|198272099|gb|EDY96368.1| putative capsid protein (F protein) [Bacteroides plebeius DSM
17135]
Length=613
Score = 227 bits (578), Expect = 6e-62, Method: Compositional matrix adjust.
Identities = 197/644 (31%), Positives = 294/644 (46%), Gaps = 66/644 (10%)
Query 5 SNIMGLHGLKNKTSRNSFDLSHRNLFTAKVGELLPCFVQEVNPGDSIKLDSSYFTRTAPL 64
+NIM + ++NK +R +DL+ + FTAK G L+P + V P D + F RT PL
Sbjct 9 ANIMSMKSVRNKPTRAGYDLTQKINFTAKAGSLIPVWWTPVLPFDDLNATVKSFVRTQPL 68
Query 65 QTAAFTRLRENVQYFFVPYQCLWKYFEGQVKNMTKN---ANGGDISQIATSPFANAKVST 121
TAAF R+R ++FVP++ +W F + M N A+G ++ N +S
Sbjct 69 NTAAFARMRGYFDFYFVPFRQMWNKFPTAITQMRTNLLHASGPVLAD-------NVPLSD 121
Query 122 EMPFISYTALHAYLNKLLNYVDSSANPTELSNPFLYNNGCWRHAESAKLLQLLGYGNFVQ 181
E+P+ + + Y+ L + N F G +R +L+ LGYG+F
Sbjct 122 ELPYFTAEQVADYIVSL----------ADSKNQF----GYYRAWLVCIILEYLGYGDFYP 167
Query 182 QFKNFSASK--PYSLLHVENAPALSVFRLLAYQKICNDFYTYRQWQPYNASLCNIDYITP 239
+ + ++ + N S F L AYQKI DF Y QW+ N S NIDYI+
Sbjct 168 YIVEAAGGEGATWATRPMLNNLKFSPFPLFAYQKIYADFNRYTQWERSNPSTFNIDYIS- 226
Query 240 DssssmdlsskfssisVSDLGKS-NMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNA 298
S +V S N+ DMR+SN D +G +P Q+G S V +S +
Sbjct 227 -----GSADSLQLDFTVEGFKDSFNLFDMRYSNWQRDLLHGTIPQAQYGEASAVPVSGSM 281
Query 299 DVYTGFDKSQWQT-LDGsafpsgsvsssnsdrsltANGKSIEHVHILPSGSITSSL---- 353
V G + T DG AF +G+V+ S L A S+ IL + S L
Sbjct 282 QVVEGPTPPAFTTGQDGVAFLNGNVTIQGSSGYLQAQ-TSVGESRILRFNNTNSGLIVEG 340
Query 354 ------SIAALRQATALQKYKEIQLANDPDFESQIEAHFGI---KPKHDMHKSRFIGGSS 404
SI ALR+A A QK+KE+ LA++ D+ SQIEAH+G K DM +++G +
Sbjct 341 DSSFGVSILALRRAEAAQKWKEVALASEEDYPSQIEAHWGQSVNKAYSDM--CQWLGSIN 398
Query 405 SMIDINPVVNQNLGAGQNQDNQAVTKAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYS 464
+ IN VVN N+ G+N + A K +G G +F +G+V+ ++ P LDY
Sbjct 399 IDLSINEVVNNNI-TGENAADIA-GKGTMSGNGSINFN-VGGQYGIVMCVFHVLPQLDYI 455
Query 465 HVGIDRTLLKTDASDFVIPELDSIGMQQTFQCELFAPTSQMTASAPDKRKYDMSRT--FG 522
T+ DF IPE D IGM+Q P P + +S FG
Sbjct 456 TSAPHFGTTLTNVLDFPIPEFDKIGMEQVPVIRGLNPVK------PKDGDFKVSPNLYFG 509
Query 523 YAPRYSEYKVSFDRYNGAFCDTLKSWVTGFNTHIF---DSDRWNDRSYFSISVPQ--LFV 577
YAP+Y +K + D+ G F +LK+W+ F+ DS + D + F
Sbjct 510 YAPQYYNWKTTLDKSMGEFRRSLKTWIIPFDDEALLAADSVDFPDNPNVEADSVKAGFFK 569
Query 578 CRPDIVKDIFALQTYHDSNDDNLYVGMVNMCYATRNLSRYGLPY 621
P ++ ++FA++ D N D + R+L GLPY
Sbjct 570 VSPSVLDNLFAVKANSDLNTDQFLCSTLFDVNVVRSLDPNGLPY 613
>gi|647452987|ref|WP_025792807.1| hypothetical protein [Prevotella histicola]
Length=584
Score = 166 bits (420), Expect = 8e-41, Method: Compositional matrix adjust.
Identities = 159/574 (28%), Positives = 242/574 (42%), Gaps = 112/574 (20%)
Query 14 KNKTSRNSFDLSHRNLFTAKVGELLPCFVQEVNPGDSIKLDSSYFTRTAPLQTAAFTRLR 73
K + +RN FDLS R +F+AK G+LLP EVNP + K RT L TA++ R++
Sbjct 5 KPRLARNGFDLSSRRIFSAKAGQLLPIGCWEVNPSEHFKFSVQDLVRTTTLNTASYARMK 64
Query 74 ENVQYFFVPYQCLWKYFE--------------GQVKNMTKNANGGDISQIATSPFANAKV 119
E +FFV Y+ LW++F+ G KN T N N ++
Sbjct 65 EYYHFFFVSYRSLWQWFDQFIVGTNNPHSALNGVKKNGTTNYN---------------QI 109
Query 120 STEMPFISYTALHAYLNKLLNYVDSSANPTELSNPFLYNNGCWRHAESAKLLQLLGYG-- 177
+ +P L KL+ + +S ++ F Y+ G +AKLL +L YG
Sbjct 110 CSSVPTFD-------LGKLITRLKTSDMDSQ---GFNYSEG------AAKLLNMLNYGVT 153
Query 178 --NFVQQFKNFSASKPYSLLHVENAPA------LSVFRLLAYQKICNDFYTYRQWQPYNA 229
+N S Y + P+ +S FRLLAYQKI NDFY + W P +
Sbjct 154 NKGKFMNLENLITSTSYLPSKDDKEPSSIYACKVSPFRLLAYQKIFNDFYRNQDWTPSDV 213
Query 230 SLCNIDYITPDssssmdlsskfssisVSDLGKSNMLDMRFSNLPLDYFNGVLPTPQFGSE 289
N+D DS+ +++ MR+ D+ + PTP + S+
Sbjct 214 RSFNVDDYADDSNLTIEPDVAL-----------KFCQMRYRPYAKDWLTSMKPTPNY-SD 261
Query 290 SVVSLSQ----NADVYTGFDKSQWQTLDGsafpsgsvsssnsdrsltANGKSIEHVHILP 345
+ +L + N +V +KS +LD
Sbjct 262 GIFNLPEYVRGNGNVILTNNKSGSVSLD-------------------------------- 289
Query 346 SGSIT-SSLSIAALRQATALQKYKE-IQLANDPDFESQIEAHFGIK-PKHDMHKSRFIGG 402
SG+++ SS S+ LR A AL K E + AN D+ SQIEAHFG K P+ + +RF+GG
Sbjct 290 SGTVSPSSFSVNDLRAAFALDKMLEATRRANGLDYASQIEAHFGFKVPESRANDARFLGG 349
Query 403 SSSMIDINPVVNQNLGAGQNQDNQAVTKAAPTGQGGAS---FKFTADTFGVVIGIYRCTP 459
+ I ++ VV+ N A + + ++ G G S +F + G+++ IY P
Sbjct 350 FDNSIVVSEVVSTNGNAASDGSHASIGDLGGKGIGSMSSGTIEFDSTEHGIIMCIYSVAP 409
Query 460 VLDYSHVGIDRTLLKTDASDFVIPELDSIGMQQTFQCELFAPTSQMTASAPDKRKYDMSR 519
+Y+ +D K F PE +G Q +L T M +++
Sbjct 410 QSEYNASYLDPFNRKLTREQFYQPEFADLGYQALIGSDLICSTLGMNEKQAGFSDIELNN 469
Query 520 T-FGYAPRYSEYKVSFDRYNGAF--CDTLKSWVT 550
GY RY+EYK + D G F +L W T
Sbjct 470 NLLGYQVRYNEYKTARDLVFGDFESGKSLSYWCT 503
>gi|517172762|ref|WP_018361580.1| hypothetical protein [Prevotella nanceiensis]
Length=568
Score = 162 bits (410), Expect = 1e-39, Method: Compositional matrix adjust.
Identities = 161/613 (26%), Positives = 256/613 (42%), Gaps = 75/613 (12%)
Query 19 RNSFDLSHRNLFTAKVGELLPCFVQEVNPGDSIKLDSSYFTRTAPLQTAAFTRLRENVQY 78
RN+FD+S R+LFTA G LLP ++ P D +++++S F RT P+ +AAF +R ++
Sbjct 18 RNAFDISQRHLFTAPAGALLPVLSLDLLPHDHVEINASDFMRTLPMNSAAFMSMRGVYEF 77
Query 79 FFVPYQCLWKYFEGQVKNMTKNANGGDISQIATSPFANAKVSTEMPFISYTALHAYLNKL 138
+FVPY+ LW F+ + M S +S K T +S+ + KL
Sbjct 78 YFVPYKQLWSGFDQFITGM---------SDYKSSFMYAFKGKTPPSCVSFD-----VQKL 123
Query 139 LNYVDSSANPTELSNPFLYNNGCWRHAESAKLLQLLGYGNFVQQFKNFSASKPYSLLHVE 198
+++ + N + + F N G +R +L LLGYG + SA PY+
Sbjct 124 VDWCKT--NTAKDIHGFDKNKGVYR------ILDLLGYGKYAN-----SAGVPYTNPTST 170
Query 199 NAPALSVFRLLAYQKICNDFYTYRQWQPYNASLCNIDYITPDssssmdlsskfssisVSD 258
+ FR LAYQKI NDFY ++ Y N+D S K ++
Sbjct 171 TMGKCTPFRGLAYQKIYNDFYRNTTYEEYQLESFNVDMF--------YGSGKVKETIPNE 222
Query 259 LGKSNMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYTGFDKSQWQTLDGsafp 318
+ +R+ N D V PTP F + N +TG GS
Sbjct 223 PWDYDWFTLRYRNAQKDLLTNVRPTPLFSIDDF-----NPQFFTG----------GSDIV 267
Query 319 sgsvsssnsdrsltANGKSIEHVHILPSG--SITSSLSIAALRQATALQKYKEIQLANDP 376
+ + I ++ +G S + +S+A +R A AL+K + +
Sbjct 268 MEKGPNVTGGTHEYRDSVVIVGKNLKENGVDSKRTMISVADIRNAFALEKLASVTMRAGK 327
Query 377 DFESQIEAHFGIKPKHDMH-KSRFIGGSSSMIDINPVVNQNLGAGQNQDNQAV------T 429
++ Q+EAHFGI + + +IGG S I + V + + + T
Sbjct 328 TYKEQMEAHFGISVEEGRDGRCTYIGGFDSNIQVGDVTQSSGTTVTGTKDTSFGGYLGRT 387
Query 430 KAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFVIPELDSIG 489
TG G +F A G+++ IY P + Y +D + K + DF +PE +++G
Sbjct 388 TGKATGSGSGHIRFDAKEHGILMCIYSLVPDVQYDSKRVDPFVQKIERGDFFVPEFENLG 447
Query 490 MQQTFQCELFAPTSQMTASAPDKRKYDMSRTFGYAPRYSEYKVSFDRYNGAFC--DTLKS 547
MQ F + + TA++ K FG+ PRYSEYK + D +G F + L
Sbjct 448 MQPLFAKNISYKYNNNTANSRIKN----LGAFGWQPRYSEYKTALDINHGQFVHQEPLSY 503
Query 548 WVTGFNTHIFDSDRWNDRSYFSISVPQLFVCRPDIVKDIFALQTYHDSNDDNLYVGMVNM 607
W R S F+IS F P + D+FA+ D ++ G
Sbjct 504 WTVA-------RARGESMSNFNIST---FKINPKWLDDVFAVNYNGTELTDQVFGGCYFN 553
Query 608 CYATRNLSRYGLP 620
++S G+P
Sbjct 554 IVKVSDMSIDGMP 566
>gi|494308783|ref|WP_007173938.1| hypothetical protein [Prevotella bergensis]
gi|270333035|gb|EFA43821.1| putative capsid protein (F protein) [Prevotella bergensis DSM
17361]
Length=553
Score = 161 bits (408), Expect = 2e-39, Method: Compositional matrix adjust.
Identities = 159/619 (26%), Positives = 258/619 (42%), Gaps = 99/619 (16%)
Query 18 SRNSFDLSHRNLFTAKVGELLPCFVQEVNPGDSIKLDSSYFTRTAPLQTAAFTRLRENVQ 77
+RN+FDLS R+LFTA G LLP ++ P D +++++ F RT P+ TAAF +R +
Sbjct 16 NRNAFDLSQRHLFTAHAGMLLPVLNLDLIPHDHVEINAQDFMRTLPMNTAAFASMRGVYE 75
Query 78 YFFVPYQCLWKYFEGQVKNMTKNANGGDIS-QIATSPFANAKVSTEMPFISYTALHAYLN 136
+FFVPY LW F+ + M + + S Q TSP ++P+ + ++ LN
Sbjct 76 FFFVPYHQLWAQFDQFITGMNDFHSSANKSIQGGTSPL-------QVPYFNVDSVFNSLN 128
Query 137 KLLNYVDSSANPTELSNPFLYNNGCWRHAESAKLLQLLGYGNFVQQFKNFSASKPYSLLH 196
S + +L F Y G +R LL LLGYG ++F +F + P ++
Sbjct 129 T--GKESGSGSTDDLQYKFKY--GAFR------LLDLLGYG---RKFDSFGTAYPDNVSG 175
Query 197 VENAPA--LSVFRLLAYQKICNDFYTYRQWQPYNASLCNIDYITPDssssmdlsskfssi 254
++N SVFR+LAY KI D+Y ++ ++ N D +
Sbjct 176 LKNNLDYNCSVFRILAYNKIYQDYYRNSNYENFDTDSFNFDKFK---------GGLVDAK 226
Query 255 sVSDLGKSNMLDMRFSNLPLDYFNGVLPTPQFGSESVVSLSQNADVYTGFDKSQWQTLDG 314
V+DL K +R+ N DYF + + F + N ++
Sbjct 227 VVADLFK-----LRYRNAQTDYFTNLRQSQLFSFTTAFEDVDNINI-------------- 267
Query 315 safpsgsvsssnsdrsltANGKSIEHVHI-LPSGSITSSLSIAALRQATALQKYKEIQLA 373
+ ++G + V+ + + S S+++LR A A+ K + +
Sbjct 268 -----------APRDYVKSDGSNFTRVNFGVDTDSSEGDFSVSSLRAAFAVDKLLSVTMR 316
Query 374 NDPDFESQIEAHFGIK-PKHDMHKSRFIGGSSSMIDINPVVNQNLGAGQNQDNQA----V 428
F+ Q+ AH+G++ P + ++GG S + ++ V + +A
Sbjct 317 AGKTFQDQMRAHYGVEIPDSRDGRVNYLGGFDSDMQVSDVTQTSGTTATEYKPEAGYLGR 376
Query 429 TKAAPTGQGGASFKFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFVIPELDSI 488
TG G F A GV++ IY P + Y +D + K D D+ PE +++
Sbjct 377 VAGKGTGSGRGRIVFDAKEHGVLMCIYSLVPQIQYDCTRLDPMVDKLDRFDYFTPEFENL 436
Query 489 GMQQTFQCELFAPTSQMTASAPDKRKYDMSRTFGYAPRYSEYKVSFDRYNGAFC--DTLK 546
GMQ + S + P + GY PRYSEYK + D +G F D L
Sbjct 437 GMQPLNSSYI----SSFCTTDP------KNPVLGYQPRYSEYKTALDVNHGQFAQSDALS 486
Query 547 SW-VTGFNTHIFDSDRWNDRSYFSISVPQL----FVCRPDIVKDIFALQTYHDSNDDNLY 601
SW V+ F RW + PQL F P + IF + +D +Y
Sbjct 487 SWSVSRFR-------RW-------TTFPQLEIADFKIDPGCLNSIFPVDYNGTEANDCVY 532
Query 602 VGMVNMCYATRNLSRYGLP 620
G ++S G+P
Sbjct 533 GGCNFNIVKVSDMSVDGMP 551
>gi|496521299|ref|WP_009229582.1| capsid protein [Prevotella sp. oral taxon 317]
gi|288330570|gb|EFC69154.1| putative capsid protein (F protein) [Prevotella sp. oral taxon
317 str. F0108]
Length=541
Score = 157 bits (396), Expect = 7e-38, Method: Compositional matrix adjust.
Identities = 147/544 (27%), Positives = 233/544 (43%), Gaps = 99/544 (18%)
Query 19 RNSFDLSHRNLFTAKVGELLPCFVQEVNPGDSIKLDSSYFTRTAPLQTAAFTRLRENVQY 78
R++FDLS ++L+TA G LLP ++ D I++ + F RT P+ +AAF +R ++
Sbjct 18 RSAFDLSQKHLYTAPAGALLPVLSVDLMFHDHIRIQAQDFMRTMPMNSAAFISMRGVYEF 77
Query 79 FFVPYQCLWKYFEGQVKNMTKNANGGDISQIATSPFANAKVSTEMPFISYTALHAYLNKL 138
FFVPY LW ++ + +M D S A K +P + ++ ++ +
Sbjct 78 FFVPYSQLWHPYDQFITSMN------DYRSSVVSSAAGDKALDSVPNVKLADMYKFVRER 131
Query 139 LNYVDSSANPTELSNPFLYNNGCWRHAESAKLLQLLGYGNFVQQFKNFSASKPYSLLHVE 198
+ D P NN C +L+ LLGYG + S+ P LL+
Sbjct 132 TD-KDIFGYPHS-------NNSC-------RLMDLLGYGKPIT-----SSKTPVPLLYTG 171
Query 199 NAPALSVFRLLAYQKICNDFYTYRQWQPYNASLCNIDYITPDssssmdlsskfssisVSD 258
N +++FRLLAY KI +D+Y ++ + NID+ K + + +D
Sbjct 172 N---VNLFRLLAYNKIYSDYYRNTTYEGVDVYSFNIDH------------KKGTFVPTAD 216
Query 259 LGKSNMLDMRFSNLPLDYFNGVLPTPQF--GSESVVSLSQNADVYTGFDKSQWQTLDGsa 316
K L++ + N PLD++ + PTP F GS+S S+ Q +D TG
Sbjct 217 EFK-KYLNLHYRNAPLDFYTNLRPTPLFTIGSDSFSSVLQLSDP-TG------------- 261
Query 317 fpsgsvsssnsdrsltANGKSIEHVHILPSGSITSSLSIAALRQATALQKYKEIQLANDP 376
+A+G S + P L+++A+R A AL K I +
Sbjct 262 -----------SAGFSADGNSAKLNMASP-----DVLNVSAIRSAFALDKLLSISMRAGK 305
Query 377 DFESQIEAHFGIKPKHDMH-KSRFIGGSSSMIDINPVVNQNLGAGQNQDNQAVTKAA--- 432
+ QIEAHFG+ + ++GG S + + V + N K A
Sbjct 306 TYAEQIEAHFGVTVSEGRDGQVYYLGGFDSNVQVGDVTQTSGTTNPNVSEVGNAKLAGYL 365
Query 433 ------PTGQGGASFKFTADTFGVVIGIYRCTPVLDYSHVGIDRTLLKTDASDFVIPELD 486
TG G +F A GV++ IY P + Y + +D + K D+ IPE +
Sbjct 366 GKITGKGTGSGYGEIQFDAKEPGVLMCIYSVVPAMQYDCMRLDPFVAKQTRGDYFIPEFE 425
Query 487 SIGMQQTFQCELFAPTSQMTASAPDKRKYDMSRTFGYAPRYSEYKVSFDRYNGAFC--DT 544
++GMQ P A D ++G+ PRYSEYK +FD +G F +
Sbjct 426 NLGMQP------IVPAFVSLNRAKDN-------SYGWQPRYSEYKTAFDINHGQFANGEP 472
Query 545 LKSW 548
L W
Sbjct 473 LSYW 476
Lambda K H a alpha
0.319 0.134 0.404 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4697304392193