bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-9_CDS_annotation_glimmer3.pl_2_1
Length=497
Score E
Sequences producing significant alignments: (Bits) Value
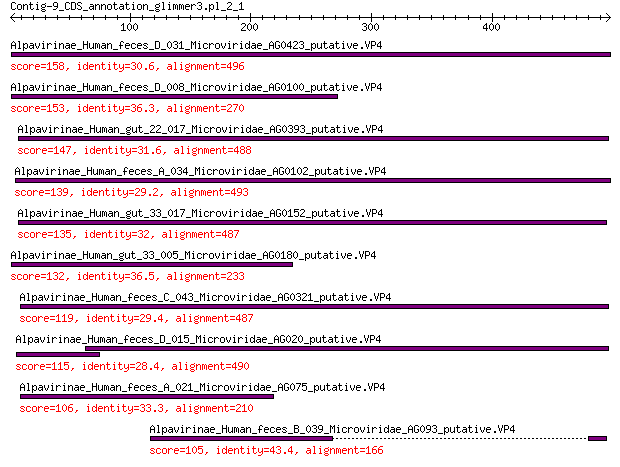
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 158 4e-44
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 153 3e-42
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 147 4e-40
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 139 2e-37
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 135 3e-36
Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4 132 3e-35
Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4 119 6e-31
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 115 2e-29
Alpavirinae_Human_feces_A_021_Microviridae_AG075_putative.VP4 106 3e-26
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 105 6e-26
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 158 bits (400), Expect = 4e-44, Method: Compositional matrix adjust.
Identities = 152/556 (27%), Positives = 231/556 (42%), Gaps = 98/556 (18%)
Query 2 IKNSFCKCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITL 61
I+ F C H + NPYT E ++VPCG C AC K+ + +S V++H F+TL
Sbjct 7 IQKVFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLVSRHVYFVTL 66
Query 62 TYANRYIPRATF-VDSLERPF---------GNDLV-------DKETGEILGPSDMKQEDI 104
TYA RYIP + +++L+ F N + K I G + +
Sbjct 67 TYAQRYIPYYEYEIEALDADFLAITAHCCDRNPMYRTYTYRGTKHKLRIRGLASPNVKSF 126
Query 105 DRLLNKFY------------------LFGDVPYLRKTDLQLFFKRLRYYVSKQCPSEKVR 146
+N+ Y L G +PYL D+ L+ KR+R Y+SK +E +
Sbjct 127 SFSVNRDYWTSYAQKANLSFNGKYPALSGRIPYLLHGDVSLYMKRVRKYISKLGINETIH 186
Query 147 YFAVGEYGPVHFRPHYHILLFLQSDEALQVCSENISQAWTLGRIDCQISKGQCSSYVASY 206
+ VGEYGP FRPH+H+LLF SDE Q S W GR+DC S+G YV+SY
Sbjct 187 TYIVGEYGPSSFRPHFHLLLFFDSDELAQNIIRIASSCWRFGRVDCSASRGDAEDYVSSY 246
Query 207 VNSSCTIP-KVFKLSSVCPFNVHSQKLGQGFLDCQREKIYSSTPQNFVKRSIVLNGK--- 262
+NS +IP + ++ ++ PF S K G F + +K S NF + +LNGK
Sbjct 247 LNSFSSIPLHIQEIRAIRPFARFSNKFGYSFFESSIKKAQSG---NFDE---ILNGKSLP 300
Query 263 YKEFDV----WRSCY--AYYFPRCKGFASKSSRERAYSYGIYDTARRLFSSSETTFSLAK 316
Y FD WR+ +Y P + + Y R F + T F
Sbjct 301 YNGFDTTIFPWRAIIDTCFYRPALRRHSDIHELTEILRYARNFKQRPAFQKA-TLFQSPG 359
Query 317 EIAFYIKHFHFTDDTYLLDLFGHVSDQKSLLDLSNYFLDRDAMVRPVESDEFNRWVHRIY 376
+ Y++ + ++ SD LS LD +++ E E + R+Y
Sbjct 360 IMYAYLQDLGPSAAAKFIE-----SDYPLFRILSFLKLDYTKIIQG-EPSEVRSFHSRLY 413
Query 377 TELLVSKHFLYFVCTHTTLAERKSKQRMIEEFYSYLDYMHLTTFFESQQEFYE------- 429
L S+ FL + +T L+ R ++Y+ + E+ FY
Sbjct 414 MFLRQSELFLNGI-GYTLLSTR-------------VEYLLIKKSLENSINFYNERERKSL 459
Query 430 SDLIGDDDLCTDQWENSYYPYFYYNVHTNEPFEKTPVYRLYASDVKKLFND--------R 481
DL D + W + ++ + + R SD L D R
Sbjct 460 QDLFHDSEAFESDWSDIFWDR-----------RQEKIKRFVDSDYGSLCRDKLHSEIRKR 508
Query 482 IKHKKLNDANKIFIDE 497
IKH+++NDA IF +
Sbjct 509 IKHREINDAVGIFTKQ 524
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 153 bits (386), Expect = 3e-42, Method: Compositional matrix adjust.
Identities = 98/307 (32%), Positives = 145/307 (47%), Gaps = 37/307 (12%)
Query 2 IKNSFCKCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITL 61
I+ F C H + NPYT E ++VPCG C AC K+ + +S +++H F+TL
Sbjct 7 IQKVFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLISRHVYFVTL 66
Query 62 TYANRYIPRATF-VDSLERPF---------GNDLV-------DKETGEILGPSDMKQEDI 104
TYA RYIP + +++L+ F N + K I G + K +
Sbjct 67 TYAQRYIPYYEYEIEALDADFLAITAHCRDRNPMYRTYTYRGTKHKLRIRGLASPKVKSF 126
Query 105 DRLLNKFY------------------LFGDVPYLRKTDLQLFFKRLRYYVSKQCPSEKVR 146
+N+ Y L +PYL D+ L+ KR+R Y+SK +E +
Sbjct 127 SFSVNRDYWTSYAQKANLSFDGKYPALSDRIPYLLHDDVSLYMKRVRKYISKLGINETIH 186
Query 147 YFAVGEYGPVHFRPHYHILLFLQSDEALQVCSENISQAWTLGRIDCQISKGQCSSYVASY 206
+ VGEYGPV FRPH+H+LLF SDE Q W GR+DC S+G YV+SY
Sbjct 187 TYVVGEYGPVTFRPHFHLLLFFNSDELAQSIVRIARSCWRFGRVDCSASRGDAEDYVSSY 246
Query 207 VNSSCTIP-KVFKLSSVCPFNVHSQKLGQGFLDCQREKIYSSTPQNFVK-RSIVLNGKYK 264
+NS +IP + ++ ++ PF S K G F + +K S + +S+ NG
Sbjct 247 LNSFSSIPLHIQEIRAIRPFARFSNKFGFSFFESSIKKAQSGDFDEILNGKSLPYNGFNT 306
Query 265 EFDVWRS 271
WR+
Sbjct 307 TIFPWRA 313
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 147 bits (370), Expect = 4e-40, Method: Compositional matrix adjust.
Identities = 154/573 (27%), Positives = 249/573 (43%), Gaps = 108/573 (19%)
Query 8 KCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRY 67
+C HP+ +VN YT+E +VV CGHC +C+L ++S S ++ F+TLTYA +
Sbjct 13 RCQHPRTVVNKYTHEPVVVSCGHCPSCILRRSSVQTNLLTTYSAQFRYVYFVTLTYAPSF 72
Query 68 IPRATF------------------VDSLERP------FGNDLVDKETGEILGPSD----- 98
+P +D L+ FG V + + L S
Sbjct 73 LPTLEVSVVETCTDDIADVSCVPNIDELDASDPNTYLFGFRSVPRSSSVKLKSSTVERTF 132
Query 99 ----------MKQEDIDRLLNKFY--LFGDVPYLRKTDLQLFFKRLR-YYVSKQCPSEKV 145
MK +++ +L K + +PY+ DL LF KRLR YY+ EK+
Sbjct 133 KDPDVKFSYPMKPKELLSILGKINHNVPNRIPYICNRDLDLFLKRLRSYYLD-----EKL 187
Query 146 RYFAVGEYGPVHFRPHYHILLFLQSDEALQVCSENISQAWTLGRIDCQISKGQCSSYVAS 205
RY+AV EYGP FRPH+H+LLF S+ + EN+S+AW+ GR D +S+G + YVAS
Sbjct 188 RYYAVSEYGPTSFRPHWHLLLFSNSERFSRTVCENVSKAWSYGRCDASLSRGFAAPYVAS 247
Query 206 YVNSSCTIPKVFKLSS--VCPFNVHSQKLGQGFLDCQREKIYSSTPQNFVKRSIVLNGKY 263
YVNS +P + V P + HS + L ++ ++ LNG
Sbjct 248 YVNSFVALPDFYTQMPKVVRPKSFHSIGFTESNLFPRKVRVAEVDEIT----DKCLNGVR 303
Query 264 KEFDVW----RSCYAY---YFPRCKG-------------FASKSSRERAYSYGIYDTARR 303
E D + + + Y FPR FA+ ++ ER G D
Sbjct 304 VERDGYFRTIKPTWPYLLRLFPRFSDAIRKSPSSIYQLLFAAFTAPERVIRSGCADIGCD 363
Query 304 LF--SSSETTFSLAKEIAFYIKHFHFTDDTYLLDLFGHVSDQKSLLDLSNYFLDRDAMVR 361
F SS ++ S K+ Y+ ++ +++ +S Q SL L +
Sbjct 364 PFGESSKQSILSFCKQYLNYVDNYGKSNEHR-----NFLSPQASLPHSDVLILSECRLYD 418
Query 362 PVESDEFNRWVHRIYTELLVSKHFL----------YFVCTHTTLAE---RKSKQRMIEE- 407
V+ + +R V R+Y L F+ F + T + R+ R+I E
Sbjct 419 GVDLETTHR-VSRVYRFFLGISKFIRTYSTDGCSELFWSSGTPGGDLFCRERFLRIISEK 477
Query 408 ---FYSYLDYMHLTTFFESQQEFYESDLIGDDDLCTDQWENSYYPYFY-YNVHTNE-PFE 462
F++ +Y L F+++ ++ + +L+ +E Y + Y +V NE P+
Sbjct 478 IVNFWNRYEYNRLVDFYQTLEDSNDKELV--------DFELRNYSFRYNRSVRDNERPYN 529
Query 463 KTPVYRLYASDVKKLFNDRIKHKKLNDANKIFI 495
+ P+ R A+ D++KHKK+ND + IF+
Sbjct 530 ELPLVRRLAAASLMKCRDKVKHKKVNDLSGIFL 562
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 139 bits (349), Expect = 2e-37, Method: Compositional matrix adjust.
Identities = 144/559 (26%), Positives = 229/559 (41%), Gaps = 99/559 (18%)
Query 5 SFCKCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYA 64
++ CLHP+ I N YT + + VPCG C+ C+ K + +C+++ +++ FITLTY+
Sbjct 8 TYGSCLHPRVIKNKYTGDPIYVPCGTCEFCVHNKAIKAELKCNVQLASSRYCEFITLTYS 67
Query 65 NRYIPRATFVD---------SLER----------------PFGNDLVDKETGEILGPSDM 99
Y+P F SL R F + D +T + +
Sbjct 68 TEYLPVGKFYQGPRGEVRFCSLPRDFVYSYKTVQGYTRNISFNDKTFDFDTQLSWSSAQL 127
Query 100 KQE----------DIDRLLNKFYLFGDVPYLRKTDLQLFFKRLRYYVSKQCPSEKVRYFA 149
Q+ D + N+ Y+ G V YL D+QLFFKRL + ++ +EK+ Y+
Sbjct 128 LQKKAHLHYTSFPDGRCIYNRPYMEGLVGYLNYHDIQLFFKRLNQNI-RRITNEKIYYYV 186
Query 150 VGEYGPVHFRPHYHILLFLQSDEALQVCSENISQAWTLGRIDCQISKGQCSSYVASYVNS 209
VGEYGP FRPH+HILLF S + + + +S++W G D Q S YVA YVNS
Sbjct 187 VGEYGPTTFRPHFHILLFHDSRKLRESIRQFVSKSWRFGDSDTQPVWSSASCYVAGYVNS 246
Query 210 SCTIPKVFKLSS-VCPF-----NVHSQKLGQGFLDCQREKIYSSTPQNFVKRSIVLNGKY 263
+ +P FK S + PF N + F + E+I+S + R + LNGK
Sbjct 247 TACLPDFFKNSRHIKPFGRFSVNFAESAFNEVFKPEENEEIFSLF---YDGRVLNLNGKP 303
Query 264 KEFDVWRSCYAYYFPRCKGFASKSSRERAYSYGIYD-----TARRLFSSSETTFSLAKEI 318
RS +PR + + + A+ F + F L+K +
Sbjct 304 TIVRPKRSHINRLYPRLDKSKHATVVDDIRIASVVSRLPQVVAKFGFIDEVSDFELSKRV 363
Query 319 AFYIKHFHFTDDTYL---------------LDLFGHVSDQKSLLDLSNYFLDRDAMVR-- 361
+ I+ H D L L L+ + SD+ + L +V+
Sbjct 364 YYLIRR-HLEVDCSLDYASDELRVIYNACRLSLYINFSDESGCAAIYRLLLQYRNLVKNW 422
Query 362 ---PVESDEFNRWVHRIYTELLVSKHFLYFVCTHTTLAERKSKQRMIEEFYSYLDYMHLT 418
PV S F +HR + H Y C+ +L ++ K +E++ +
Sbjct 423 ITAPVGSVAFTGQLHRAVRAI----HSFYDYCSRRSLHDQLVK---VEKWSN-------D 468
Query 419 TFFESQQEFYESDLIGDDDLCTDQWENSYYPYFYYNVHTNEPFEKTPVYRLYASDVKKLF 478
++ + Y + D D+ + +E + V R +D
Sbjct 469 SYVRANLSIYYFYPLTDVDIMMKSF--------------SEIVFDSQVLRASYADYAADN 514
Query 479 NDRIKHKKLNDANKIFIDE 497
+RIKHK LND N + I E
Sbjct 515 RERIKHKALNDKNSLLIAE 533
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 135 bits (340), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 156/574 (27%), Positives = 246/574 (43%), Gaps = 112/574 (20%)
Query 8 KCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDL-ESYVA--KHTLFITLTYA 64
+C +P+ +VN YT+E +VV CG C +C+L R Q +L +Y A ++ F+TLTYA
Sbjct 13 RCQNPRTVVNKYTHEPVVVSCGACPSCVL---RRSGIQTNLLTTYSAQFRYVYFVTLTYA 69
Query 65 ------------------------------------NRY------IPRATFV----DSLE 78
NRY +PR+ V ++E
Sbjct 70 PCFLPTLEVSVIETCTDDIADVSSVPDINDLDPCDNNRYLFGFCSVPRSASVKLKNSTVE 129
Query 79 RPFGNDLVDKETGEILGPSDMKQEDIDRLLNKFY--LFGDVPYLRKTDLQLFFKRLRYYV 136
R F + E+ MK +++ +L+K + +PY+ DL LF KRLR Y
Sbjct 130 RTFKDP-------EVRFSYPMKPKELLSILDKINHNVPNRIPYICNRDLDLFLKRLRSYY 182
Query 137 SKQCPSEKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVCSENISQAWTLGRIDCQISK 196
P EK+RY+AV EYGP +RPH+H+LLF S++ + EN+S+AW+ GR D +S+
Sbjct 183 ----PYEKLRYYAVSEYGPTSYRPHWHLLLFSNSEQFSKTILENVSKAWSYGRCDASLSR 238
Query 197 GQCSSYVASYVNSSCTIPKVFKLSS--VCPFNVHSQKLGQGFLDCQREKIYSSTPQNFVK 254
G + YVASYVNS +P + + P + HS + L ++ +I S
Sbjct 239 GFAAPYVASYVNSFVALPSFYTEMPRFLRPKSFHSIGFTESNLFPRKVRI--SEIDEVTD 296
Query 255 RSIVLNGKYKE----FDVWRSCYAY---YFPRCKG-------------FASKSSRERAYS 294
+ LNG E F + + + Y FPR FA+ ++ R
Sbjct 297 K--CLNGVRVERDGYFRILKPSWPYLLRLFPRFSDAIRKSPSSIYQLLFAAFTAPARVIR 354
Query 295 YGIYDTARRLF--SSSETTFSLAKEIAFYIKHFHFTDDTYLLDLFGHV----SDQKSLLD 348
G D F +S ++ S K+ Y+ + H + + L H SD L +
Sbjct 355 SGCADIGCDPFVENSKQSLLSFCKQYLNYVDN-HGKSNEFKNFLSPHADLPHSDVLILTE 413
Query 349 LSNY-FLDRDAMVRPVESDEFNRWVH---RIYTELLVSKHFLYFVCTHTTLAERKSKQRM 404
Y +D +A R F + R Y+ S+ F L R+ R+
Sbjct 414 CRLYDGVDLEAAHRLSRVYRFFLGIAKFIRTYSTDGCSELFWSSGTPGGELFGRERFLRI 473
Query 405 IEE----FYSYLDYMHLTTFFESQQEFYESDLIGDDDLCTDQWENSYYPYFYYNVHTNEP 460
I E F++ DYM L F+++ ++ D +L + N + Y +P
Sbjct 474 ISEKIVVFWNRYDYMRLVDFYQTLED------SNDKELVDFEIRNYSFCYNRSVRDKEKP 527
Query 461 FEKTPVYRLYASDVKKLFNDRIKHKKLNDANKIF 494
+ + P+ R A+ D++KHKK+ND IF
Sbjct 528 YHELPLVRRLAAASLMKCRDKVKHKKINDMFGIF 561
> Alpavirinae_Human_gut_33_005_Microviridae_AG0180_putative.VP4
Length=536
Score = 132 bits (331), Expect = 3e-35, Method: Compositional matrix adjust.
Identities = 85/271 (31%), Positives = 130/271 (48%), Gaps = 42/271 (15%)
Query 2 IKNSFCKCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITL 61
+ +C CLHP I N YT + + V CG C+ C+ K + +C+++ ++ F+TL
Sbjct 5 VVKPYCSCLHPVIIKNKYTGDPIYVECGTCEVCLSNKAIQKELRCNIQLASSRCCFFVTL 64
Query 62 TYANRYIPRA--------------------TFVDSLERPFGNDLVDKETGEILGPSDMKQ 101
TYA +IP A T+V S D+E P++++
Sbjct 65 TYATEHIPVARFYKLNDSYHLCCVPRDHVYTYVTSQGYNRKMSFCDEEFDY---PTNLRD 121
Query 102 EDIDRLLNKFYL----------------FGDV-PYLRKTDLQLFFKRLRYYVSKQCPSEK 144
E + LL+K +L GD+ PYL D+QLF KR+ + K+ EK
Sbjct 122 EAVTALLDKTHLDRTVYPDGRSVVKYPNMGDLLPYLNYRDVQLFHKRINQQI-KKYTDEK 180
Query 145 VRYFAVGEYGPVHFRPHYHILLFLQSDEALQVCSENISQAWTLGRIDCQISKGQCSSYVA 204
+ + VGEYGP FRPH+H+L F S+ QV + + +AW G D Q SSYVA
Sbjct 181 IYSYTVGEYGPKTFRPHFHLLFFFDSERLAQVFGQLVDKAWRFGNSDTQRVWSSASSYVA 240
Query 205 SYVNSSCTIPKVFKLS-SVCPFNVHSQKLGQ 234
Y+NSS +P+ ++ + + PF SQ +
Sbjct 241 GYLNSSHCLPEFYRCNRKIAPFGRFSQYFAE 271
> Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4
Length=546
Score = 119 bits (299), Expect = 6e-31, Method: Compositional matrix adjust.
Identities = 143/541 (26%), Positives = 219/541 (40%), Gaps = 83/541 (15%)
Query 9 CLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRYI 68
CLH K + N YT + M CG C+AC+ S + + +++ ++K+ +F+TLTY ++
Sbjct 15 CLHGKNVYNKYTGQMMYQSCGKCEACLSRLASARSIKVGVQASLSKYVMFVTLTYDTYHV 74
Query 69 PRATF------------------------VDSLERPFGNDLVDKETGEILGPSD-----M 99
P+ D +R G D E D
Sbjct 75 PKCKIYSNGDNNYVLVVKPRVKDYFYYETSDGQKRLKGLSYDDDFRVEFKADKDYIENFK 134
Query 100 KQEDIDRLLNKFYLFGDVPYLRKTDLQLFFKRLRYYVSKQCPSEKVRYFAVGEYGPVHFR 159
KQ ++D +L YL + D QLF KR+R + + EK+ + VGEY P HFR
Sbjct 135 KQANLDVKGCYPHLKDMYGYLSRKDCQLFMKRVRKQI-RNYTDEKIHTYIVGEYSPKHFR 193
Query 160 PHYHILLFLQSDEALQVCSENISQAWTLGRIDCQISKGQCSSYVASYVNSSCTIPKVFKL 219
PH+HIL F S+E Q + W G +D S+G SYVA YVNS +P + L
Sbjct 194 PHFHILFFFNSNELSQSFGSIVRSCWKFGLVDWSQSRGDAESYVAGYVNSFARLP--YHL 251
Query 220 SS---VCPFNVHSQKLGQGFLDCQREKIYSS-----TP--QNFVKR-SIVLNGKYKEFDV 268
S + PF S + D ++ + S TP F+ + ++NGK
Sbjct 252 GSSPKIAPFGRFSNHFSESCFDDAKQTLRDSFFGQKTPVFAPFLNGVTNLVNGKLLLTRP 311
Query 269 WRSCYAYYFPRCKGFASKSSRE-----RAYSYGIYDTARRLFSSSETTFSLAKEIAFYI- 322
RSC F R SS E RA + + + +R F+ ++ F +
Sbjct 312 SRSCVDSCFFRKARDGRLSSHELLHLIRAVCH-VVEQGKRFFARDGLNYTFLNHARFIVK 370
Query 323 --KHFHFTDDTYLLDLFGHVSDQKSLLDLSNYFLDRDAM-VRPVESDEFNRWVHRIYTEL 379
K F + LL L S +LL S L +D + + DE IY L
Sbjct 371 ACKSFRYQKREKLLSL---PSRLVTLLYYSRVDLTKDIQDYKNPDVDE--AMAQSIYRVL 425
Query 380 LVSKHFLYF-VCTHTTLAERKSKQRMIEEFYSYLDYMHLTTFFESQQEFYESDLIGDDDL 438
L + F+ F E M E+Y LDY +L F+ ++ E DL+
Sbjct 426 LYTMRFVRFWKIDKMPYHEGIRLLDMCREYYRLLDYQYLRQRFQFLEQC-EEDLLD---- 480
Query 439 CTDQWENSYYPYFYYNVHTNEPFE----KTPVYRLYASDVKKLFNDRIKHKKLNDANKIF 494
FY +H E F +T V + +++ + IKH+++ND N +
Sbjct 481 ------------FY--LHPTENFADYLPRTIVTGINQYSLER-YQKAIKHREINDLNLQY 525
Query 495 I 495
+
Sbjct 526 V 526
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 115 bits (288), Expect = 2e-29, Method: Compositional matrix adjust.
Identities = 117/442 (26%), Positives = 195/442 (44%), Gaps = 42/442 (10%)
Query 63 YANRYIPRATFVDSLERPFGNDLV--DKETGEILGPSDMKQEDIDRLLNKFYLFGDVPYL 120
Y R IPR + + + + + +LV E E L + D + +F G + Y+
Sbjct 155 YILRSIPRVSKLQNFKDEYFEELVWMLPEIAESLKKKN--NTDANGAFPQFK--GLLKYV 210
Query 121 RKTDLQLFFKRLRYYVSKQCPS-EKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQVCSE 179
D QLF KRLR Y+SK+ EK+ + V EY P FRPH+HIL F SDE + +
Sbjct 211 NIRDYQLFAKRLRKYLSKKVGKYEKIHSYVVSEYSPKTFRPHFHILFFFDSDEIAKNFRQ 270
Query 180 NISQAWTLGRIDCQISKGQCSSYVASYVNSSCTIPKVFKL-SSVCPFNVHSQKLGQGFLD 238
+ Q+W LGR+D Q+++ Q +SYV++Y+NS +IP V+K S+ P + S G +
Sbjct 271 AVYQSWRLGRVDTQLAREQANSYVSNYLNSVVSIPFVYKAKKSIRPRSRFSNLFGFEEVK 330
Query 239 CQREKIYSSTPQNFVKRSIVLNGKYKEFDVWRSCYAYYFPRCKGFASKSSRERAYSYGIY 298
K F S + N K+ + S FPR + R + YG+
Sbjct 331 EGIRKAQDKRAALFDGLSYISNQKFVRYVPSGSLIDRLFPRFTYYDGSFLRRSSQIYGVV 390
Query 299 DTARRLFSSS----ETTFSLAKEIAFYIKHFHFTDDTYLLDLFGHVSDQKSLLDLSNYFL 354
RLF+ + E T E + ++F + D ++ + ++ L
Sbjct 391 QQILRLFARNEPFEEATPGNVSEFICWWCAYNFRQGCQIKDFPDYMQEFLHIVR-----L 445
Query 355 DRDAMVR-PVESDEFNRWVHRIYTELLVSKHFLYFVCTHTTLAERKSKQRMIEEFYSYLD 413
DR++ + + + +R+++R F +SK + +E FY Y D
Sbjct 446 DRESFLNWDIPKGKISRFLYR-------------FNMFEKMKGSFRSKLKAVELFYDYRD 492
Query 414 YMHLTTFFESQQEFYESDLIGDDDLCTDQWENSYYPYFYYNVHTNEPFEKTPVYRLYASD 473
Y L + + Y L+ + +D+ +S+Y V N EK + ++
Sbjct 493 YQSL------KNQLYMQQLVFSELGYSDELLDSFYVKPNLKVLKNIYTEK-----WHDTN 541
Query 474 VKKLFNDRIKHKKLNDANKIFI 495
++ R+KHK LND N +F+
Sbjct 542 YHEVHYFRVKHKVLNDQNDVFL 563
Score = 60.5 bits (145), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 25/69 (36%), Positives = 39/69 (57%), Gaps = 0/69 (0%)
Query 6 FCKCLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYAN 65
F +C HP+ I N YT + + V CG C C++ K R +CD Y ++ F+TLTY
Sbjct 9 FNRCEHPQVIQNKYTGDYVKVDCGQCPYCLIKKADRSTQKCDFVKYNHRYCYFVTLTYNT 68
Query 66 RYIPRATFV 74
+Y+P+ +
Sbjct 69 QYVPKMSLT 77
> Alpavirinae_Human_feces_A_021_Microviridae_AG075_putative.VP4
Length=547
Score = 106 bits (264), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 70/249 (28%), Positives = 113/249 (45%), Gaps = 39/249 (16%)
Query 9 CLHPKKIVNPYTNESMVVPCGHCQACMLAKNSRYAFQCDLESYVAKHTLFITLTYANRYI 68
C P+ + ++++ + + C C C+L K+ + + K+ F+ L YA +I
Sbjct 16 CNQPRYLRQLHSDDVLSIGCNECLPCLLKKSRNDTLDVTISTQTFKYIFFVNLDYATPFI 75
Query 69 P----------RATFVDSLERPFGNDLVDK------------------ETGEILGPSDMK 100
P + + +S+ RP +K ET + +
Sbjct 76 PIMRHEYIPERKVIYCESMVRPSVKIPTNKRRKDGSVVFTTFSFRAADETRPFCFTAPCE 135
Query 101 QEDIDRLL---------NKFYLFGDV-PYLRKTDLQLFFKRLR-YYVSKQCPSEKVRYFA 149
E+ RL K+ ++ D PYL + D+ LF KRLR ++ K S + +
Sbjct 136 SEEEYRLFVRKVNLTANGKYPMYKDCFPYLSRYDVALFMKRLRNLFLKKYGQSIYMHSYI 195
Query 150 VGEYGPVHFRPHYHILLFLQSDEALQVCSENISQAWTLGRIDCQISKGQCSSYVASYVNS 209
VGEYGPVHFRPHYHI++ + + +AW G +I G C+SYVASYVNS
Sbjct 196 VGEYGPVHFRPHYHIIISSNDPRFAESLEHFVDKAWFFGGASAEIPDGNCASYVASYVNS 255
Query 210 SCTIPKVFK 218
S ++P +F+
Sbjct 256 SVSLPVLFR 264
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 105 bits (261), Expect = 6e-26, Method: Compositional matrix adjust.
Identities = 60/154 (39%), Positives = 87/154 (56%), Gaps = 6/154 (4%)
Query 117 VPYLRKTDLQLFFKRLRYYVSKQCPS-EKVRYFAVGEYGPVHFRPHYHILLFLQSDEALQ 175
+P+L D+Q + KRLR Y+ +Q E ++AVGEYGPVHFRPHYH+LLF SD+ +
Sbjct 180 IPFLNYVDVQNYIKRLRKYLFQQLGKYETFHFYAVGEYGPVHFRPHYHLLLFTNSDKVSE 239
Query 176 VCSENISQAWTLGRIDCQISKGQCSSYVASYVNSSCTIPKVFKLSSVCPFNVHSQKLGQG 235
V ++W LGR D Q S G SYVASYVNS C+ P +++ C + G
Sbjct 240 VLRYCHDKSWKLGRSDFQRSAGGAGSYVASYVNSLCSAPLLYR---SCRAFRPKSRASVG 296
Query 236 FLDCQREKIYSSTPQNFVKRSI--VLNGKYKEFD 267
F + + + P +++ I V+NG+ F+
Sbjct 297 FFEKGCDFVEDDDPYAQIEQKIDSVVNGRVYNFN 330
Score = 29.3 bits (64), Expect = 0.075, Method: Compositional matrix adjust.
Identities = 12/15 (80%), Positives = 13/15 (87%), Gaps = 0/15 (0%)
Query 480 DRIKHKKLNDANKIF 494
D IKHK+LNDAN IF
Sbjct 541 DMIKHKRLNDANNIF 555
Lambda K H a alpha
0.324 0.138 0.429 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 45067425