bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
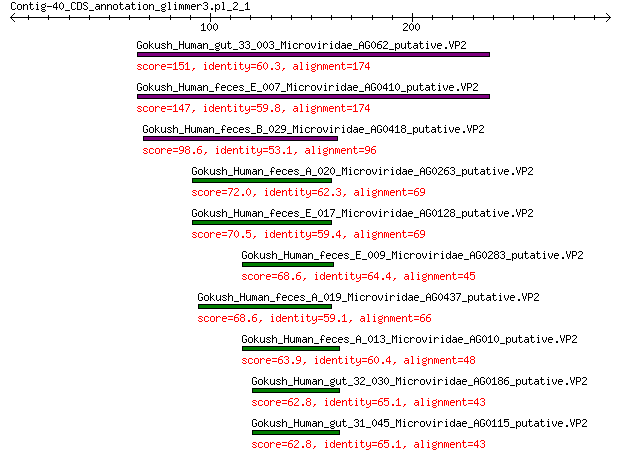
Query= Contig-40_CDS_annotation_glimmer3.pl_2_1
Length=297
Score E
Sequences producing significant alignments: (Bits) Value
Gokush_Human_gut_33_003_Microviridae_AG062_putative.VP2 151 1e-44
Gokush_Human_feces_E_007_Microviridae_AG0410_putative.VP2 147 1e-43
Gokush_Human_feces_B_029_Microviridae_AG0418_putative.VP2 98.6 9e-26
Gokush_Human_feces_A_020_Microviridae_AG0263_putative.VP2 72.0 2e-16
Gokush_Human_feces_E_017_Microviridae_AG0128_putative.VP2 70.5 8e-16
Gokush_Human_feces_E_009_Microviridae_AG0283_putative.VP2 68.6 4e-15
Gokush_Human_feces_A_019_Microviridae_AG0437_putative.VP2 68.6 4e-15
Gokush_Human_feces_A_013_Microviridae_AG010_putative.VP2 63.9 1e-13
Gokush_Human_gut_32_030_Microviridae_AG0186_putative.VP2 62.8 4e-13
Gokush_Human_gut_31_045_Microviridae_AG0115_putative.VP2 62.8 4e-13
> Gokush_Human_gut_33_003_Microviridae_AG062_putative.VP2
Length=295
Score = 151 bits (381), Expect = 1e-44, Method: Compositional matrix adjust.
Identities = 105/174 (60%), Positives = 130/174 (75%), Gaps = 0/174 (0%)
Query 64 TNDEQVMQYLKGAYQYQNAEGQRQSQFNQRSMLEQMGYNTLGAIAQGIYNHIENSAAMNF 123
TNDEQ+M+YL Y +Q + QS+ N+++ML QMGYNTLGAI QGIYNHIE +AAMN+
Sbjct 70 TNDEQIMKYLDRFYAWQGGQNAFQSKTNRQNMLMQMGYNTLGAIQQGIYNHIEQNAAMNY 129
Query 124 NSTEAMKNREWQEHMSNTAYQRAVADMKEAGLNPILAFQNggastpggsagtisgasiga 183
NS EA+ NR +QE MS+T+YQRAV DM++AGLNPILAF NGGASTPGGS TI+GAS+G
Sbjct 130 NSAEALANRNFQERMSSTSYQRAVEDMRKAGLNPILAFANGGASTPGGSGATITGASMGM 189
Query 184 psssaLGVSRASGFVPNsysseswsqsDWYNAAQSWNQMLSSTGMTPLGLQETL 237
PSSSALGVS +G VP S S S S + WY A++ LS++ TP L + L
Sbjct 190 PSSSALGVSTMNGNVPTSNYSRSESNAQWYQLAEAVGSQLSTSHSTPKALVDDL 243
> Gokush_Human_feces_E_007_Microviridae_AG0410_putative.VP2
Length=270
Score = 147 bits (372), Expect = 1e-43, Method: Compositional matrix adjust.
Identities = 104/174 (60%), Positives = 132/174 (76%), Gaps = 0/174 (0%)
Query 64 TNDEQVMQYLKGAYQYQNAEGQRQSQFNQRSMLEQMGYNTLGAIAQGIYNHIENSAAMNF 123
TND+Q+M YL YQ+Q + QS+ N+++ML QMGYNTL AI QGIYNHIEN+AAM +
Sbjct 67 TNDKQIMDYLNRYYQWQGGQNAFQSKTNRQNMLMQMGYNTLSAIQQGIYNHIENNAAMQY 126
Query 124 NSTEAMKNREWQEHMSNTAYQRAVADMKEAGLNPILAFQNggastpggsagtisgasiga 183
NS EA+ NR++QE MS+TAYQRAV DM++AGLNPILA+ GGASTPGGS TI+GAS+G
Sbjct 127 NSAEALANRQFQERMSSTAYQRAVEDMRKAGLNPILAYAQGGASTPGGSGATITGASMGM 186
Query 184 psssaLGVSRASGFVPNsysseswsqsDWYNAAQSWNQMLSSTGMTPLGLQETL 237
P+SSALGVS SG VPNSY + S S+S WY A++ +S+ +P+ L E L
Sbjct 187 PTSSALGVSTLSGNVPNSYFNRSESKSQWYQLAEAVGSQMSTGYSSPVQLTEDL 240
> Gokush_Human_feces_B_029_Microviridae_AG0418_putative.VP2
Length=272
Score = 98.6 bits (244), Expect = 9e-26, Method: Compositional matrix adjust.
Identities = 51/96 (53%), Positives = 68/96 (71%), Gaps = 3/96 (3%)
Query 67 EQVMQYLKGAYQYQNAEGQRQSQFNQRSMLEQMGYNTLGAIAQGIYNHIENSAAMNFNST 126
+ + +Y G Q Q A+G + Q N+ S++ +G NTLGAI QG+YN I+ AAM++NS
Sbjct 72 KDLAKYFLG--QSQQAQGMQSLQNNKNSLMA-LGLNTLGAIQQGVYNRIQQDAAMSYNSA 128
Query 127 EAMKNREWQEHMSNTAYQRAVADMKEAGLNPILAFQ 162
EA NR WQE MSNT+YQRA DM++AG+NPILA Q
Sbjct 129 EAAANRAWQERMSNTSYQRATEDMRKAGINPILAAQ 164
> Gokush_Human_feces_A_020_Microviridae_AG0263_putative.VP2
Length=300
Score = 72.0 bits (175), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 43/80 (54%), Positives = 51/80 (64%), Gaps = 11/80 (14%)
Query 91 NQRSMLEQMGYNTLGAIAQGIYNHIE-------NSA----AMNFNSTEAMKNREWQEHMS 139
N SM Q YN G+ QG+ N + N+A AM +NS EA NREWQEHMS
Sbjct 93 NLGSMSAQGIYNMAGSGFQGLLNSMMMNKQGSMNAALMREAMAYNSAEAALNREWQEHMS 152
Query 140 NTAYQRAVADMKEAGLNPIL 159
+TAYQRAVADM+ AG+NPIL
Sbjct 153 STAYQRAVADMRAAGINPIL 172
> Gokush_Human_feces_E_017_Microviridae_AG0128_putative.VP2
Length=300
Score = 70.5 bits (171), Expect = 8e-16, Method: Compositional matrix adjust.
Identities = 41/80 (51%), Positives = 49/80 (61%), Gaps = 11/80 (14%)
Query 91 NQRSMLEQMGYNTLGAIAQGIYNHIENSA-----------AMNFNSTEAMKNREWQEHMS 139
N SM Q YN G+ QG+ N + + AM +NS EA NREWQEHMS
Sbjct 93 NLGSMSAQGIYNMAGSGFQGLLNSMMMNKQGRMNADLMREAMAYNSAEAALNREWQEHMS 152
Query 140 NTAYQRAVADMKEAGLNPIL 159
+TAYQRAVADM+ AG+NPIL
Sbjct 153 STAYQRAVADMRAAGINPIL 172
> Gokush_Human_feces_E_009_Microviridae_AG0283_putative.VP2
Length=294
Score = 68.6 bits (166), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 29/45 (64%), Positives = 38/45 (84%), Gaps = 0/45 (0%)
Query 116 ENSAAMNFNSTEAMKNREWQEHMSNTAYQRAVADMKEAGLNPILA 160
+N AM FN+ EA KNR+WQE+MSNTA+QR +AD+K AGLNP+L+
Sbjct 64 QNRKAMEFNAAEAAKNRDWQEYMSNTAHQREIADLKAAGLNPVLS 108
> Gokush_Human_feces_A_019_Microviridae_AG0437_putative.VP2
Length=300
Score = 68.6 bits (166), Expect = 4e-15, Method: Compositional matrix adjust.
Identities = 39/77 (51%), Positives = 48/77 (62%), Gaps = 11/77 (14%)
Query 94 SMLEQMGYNTLGAIAQGIYNHIENSA-----------AMNFNSTEAMKNREWQEHMSNTA 142
SM Q YN G+ QG+ N + + AM +NS EA NREWQE+MS+TA
Sbjct 96 SMSAQGIYNMAGSGFQGLLNSMMMNKQGRMNAELMREAMAYNSAEAAHNREWQEYMSSTA 155
Query 143 YQRAVADMKEAGLNPIL 159
YQRAVADM+ AG+NPIL
Sbjct 156 YQRAVADMRAAGINPIL 172
> Gokush_Human_feces_A_013_Microviridae_AG010_putative.VP2
Length=276
Score = 63.9 bits (154), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 29/48 (60%), Positives = 36/48 (75%), Gaps = 0/48 (0%)
Query 116 ENSAAMNFNSTEAMKNREWQEHMSNTAYQRAVADMKEAGLNPILAFQN 163
+N+ AM FN+ EA KNR WQE MSNTA+QR V D+ AGLNP+L+ N
Sbjct 42 QNAKAMQFNAEEAAKNRSWQEFMSNTAHQREVRDLMAAGLNPVLSAMN 89
> Gokush_Human_gut_32_030_Microviridae_AG0186_putative.VP2
Length=289
Score = 62.8 bits (151), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 28/43 (65%), Positives = 33/43 (77%), Gaps = 0/43 (0%)
Query 121 MNFNSTEAMKNREWQEHMSNTAYQRAVADMKEAGLNPILAFQN 163
M FNS EA KNR+WQE MSNTA+QR V D+ AGLNP+L+ N
Sbjct 61 MEFNSAEAAKNRQWQEMMSNTAHQREVRDLMAAGLNPVLSAMN 103
> Gokush_Human_gut_31_045_Microviridae_AG0115_putative.VP2
Length=289
Score = 62.8 bits (151), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 28/43 (65%), Positives = 33/43 (77%), Gaps = 0/43 (0%)
Query 121 MNFNSTEAMKNREWQEHMSNTAYQRAVADMKEAGLNPILAFQN 163
M FNS EA KNR+WQE MSNTA+QR V D+ AGLNP+L+ N
Sbjct 61 MEFNSAEAAKNRQWQEMMSNTAHQREVRDLMAAGLNPVLSAMN 103
Lambda K H a alpha
0.313 0.126 0.365 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 24810089