bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-37_CDS_annotation_glimmer3.pl_2_2
Length=179
Score E
Sequences producing significant alignments: (Bits) Value
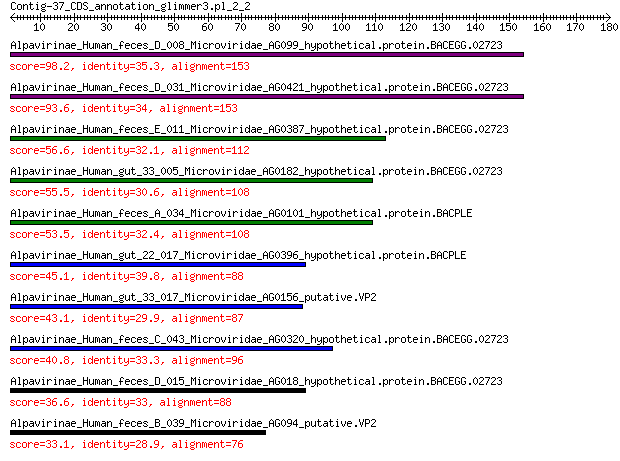
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 98.2 3e-26
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 93.6 2e-24
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 56.6 1e-11
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 55.5 2e-11
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 53.5 1e-10
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 45.1 9e-08
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 43.1 4e-07
Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.... 40.8 2e-06
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 36.6 6e-05
Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2 33.1 0.001
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 98.2 bits (243), Expect = 3e-26, Method: Compositional matrix adjust.
Identities = 54/153 (35%), Positives = 88/153 (58%), Gaps = 0/153 (0%)
Query 1 LQGKAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLN 60
LQ + Q I+NK++PAQQQ + + +AN F+QY G LS A +K ++ + A+T G
Sbjct 233 LQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSEAQVKTQIKQQALLEAQTVGQK 292
Query 61 ISNEQSEKLSEYFIRAMKEEYQANAAYYRSYKSIAGAVATGRGNLDIYDSQLSRLQRDMQ 120
++N +E+L++Y +AM EY+ANAAYY + + A + Y+S +R+ DM
Sbjct 293 LNNRLAERLADYQFKAMAAEYRANAAYYNGFYNDAWQAGMSKAAQARYESNAARIAADMS 352
Query 121 ELFASRERYSMMNRKAYYRAQQIWRDFIGGIMS 153
E+F RE+ S N YY ++ + +G + S
Sbjct 353 EIFKGREKSSWKNNPIYYNIGELLKGILGSVGS 385
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 93.6 bits (231), Expect = 2e-24, Method: Compositional matrix adjust.
Identities = 52/153 (34%), Positives = 86/153 (56%), Gaps = 0/153 (0%)
Query 1 LQGKAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLN 60
LQ + Q I+NK++PAQQQ + + +AN F+QY G LS A +K ++ + A+ G
Sbjct 233 LQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSEAQVKTQIKQQALLEAQAAGQK 292
Query 61 ISNEQSEKLSEYFIRAMKEEYQANAAYYRSYKSIAGAVATGRGNLDIYDSQLSRLQRDMQ 120
++N +E+L++Y +AM EY+ANAAYY + + A + Y+S +R+ M
Sbjct 293 LNNRLAERLADYQFKAMAAEYRANAAYYNGFYNDAWQAGMSKAAQVRYESNAARIAAQMS 352
Query 121 ELFASRERYSMMNRKAYYRAQQIWRDFIGGIMS 153
E+F RE+ S N YY ++ + +G + S
Sbjct 353 EIFKDREKASWKNNPIYYNISELLKGLLGSVGS 385
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 56.6 bits (135), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 36/112 (32%), Positives = 56/112 (50%), Gaps = 0/112 (0%)
Query 1 LQGKAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLN 60
L +A+ I+NKYL Q +QL + F +A G LSL K + + ++A T G
Sbjct 255 LSNQAKGIINKYLDTSQSLQLKLMANQSFQAFASGRLSLQQCKTEVTKQLMNMAETEGKK 314
Query 61 ISNEQSEKLSEYFIRAMKEEYQANAAYYRSYKSIAGAVATGRGNLDIYDSQL 112
ISN+ + + ++ I A++ +Y A+ Y R Y A RG D+ QL
Sbjct 315 ISNKIASETADQLIGALQWQYSADEMYSRGYAGYAREAGKSRGKGDVAKGQL 366
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 55.5 bits (132), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 33/108 (31%), Positives = 59/108 (55%), Gaps = 0/108 (0%)
Query 1 LQGKAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLN 60
L+ AQE++N YLP ++++QL A ++ +G++S KN +A E ART+G +
Sbjct 185 LRNDAQEVLNMYLPEEKRIQLQMNGAQYWNMIREGVISEEQAKNLIASRLEIEARTQGQH 244
Query 61 ISNEQSEKLSEYFIRAMKEEYQANAAYYRSYKSIAGAVATGRGNLDIY 108
ISN+ ++ ++ I A + + AA+ R Y + V G +D +
Sbjct 245 ISNKIAKSTADSIIDATRTAKENEAAFNRGYSQFSNDVGFRTGKMDRW 292
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 53.5 bits (127), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 53/108 (49%), Gaps = 0/108 (0%)
Query 1 LQGKAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLN 60
L +AQ++MN YLP ++Q+QL + A ++ G + KN LA E ART G +
Sbjct 185 LSAEAQQVMNMYLPQEKQIQLSTLGAQYWNMIRDGSIKEEQAKNLLATRLEIEARTAGQH 244
Query 61 ISNEQSEKLSEYFIRAMKEEYQANAAYYRSYKSIAGAVATGRGNLDIY 108
ISN+ + ++ I A AAY R Y + V G +D +
Sbjct 245 ISNKVARSTADSIIDATNTAKMNEAAYNRGYSQFSNDVGYRTGKMDRW 292
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 45.1 bits (105), Expect = 9e-08, Method: Compositional matrix adjust.
Identities = 35/88 (40%), Positives = 53/88 (60%), Gaps = 0/88 (0%)
Query 1 LQGKAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLN 60
L +A+ I+NKYL QQ+ L +A+ + + A G +S A K LAE + ARTRG N
Sbjct 212 LDNEAKGILNKYLDQHQQLDLSVKAADYYQRMAAGYVSYAEAKKALAEEALAAARTRGQN 271
Query 61 ISNEQSEKLSEYFIRAMKEEYQANAAYY 88
ISNE + +++E I A +++AAY+
Sbjct 272 ISNEVASRIAESQIAANIAANESSAAYH 299
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 43.1 bits (100), Expect = 4e-07, Method: Compositional matrix adjust.
Identities = 26/87 (30%), Positives = 49/87 (56%), Gaps = 0/87 (0%)
Query 1 LQGKAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLN 60
L+ A+ ++N+YL QQQ L ++ ++Q +QG L+ K +A+ + AR +G
Sbjct 212 LEADAKTVLNRYLDQQQQADLNVKASVYYNQMSQGHLNYNQAKKVIADEILTYARIKGQK 271
Query 61 ISNEQSEKLSEYFIRAMKEEYQANAAY 87
+SN+ +E ++ IRA ++NA +
Sbjct 272 LSNKVAEATADSLIRATNAANRSNAEF 298
> Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.protein.BACEGG.02723
Length=396
Score = 40.8 bits (94), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 32/96 (33%), Positives = 46/96 (48%), Gaps = 5/96 (5%)
Query 1 LQGKAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLN 60
L AQ +NKYL QQ+ L + A S A L+ A + +A ++LA G
Sbjct 209 LTADAQRTLNKYLDMGQQLSLITKMAEYSSITAGTELTKAKYRTEIANEIKTLAEANGQK 268
Query 61 ISNEQSEKLSEYFIRAMKEEYQANAAYYRSYKSIAG 96
ISNE + ++ I AM +E + YRSY + G
Sbjct 269 ISNEIARSTAQSLIDAMNKENE-----YRSYDAALG 299
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 36.6 bits (83), Expect = 6e-05, Method: Compositional matrix adjust.
Identities = 29/88 (33%), Positives = 44/88 (50%), Gaps = 6/88 (7%)
Query 1 LQGKAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLN 60
L +++ I+NKYL QQQ L +A+ +G L + + L+ + AR RGLN
Sbjct 239 LDAESKTILNKYLDQQQQADLNVKAAHYEELINRGQLHVVEARELLSREVLNYARARGLN 298
Query 61 ISNEQSEKLSEYFIRAMKEEYQANAAYY 88
ISN + K ++ + Y NAA Y
Sbjct 299 ISNWVAAKSAKGLV------YANNAANY 320
> Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2
Length=380
Score = 33.1 bits (74), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/76 (29%), Positives = 36/76 (47%), Gaps = 0/76 (0%)
Query 1 LQGKAQEIMNKYLPAQQQMQLYSYSANLFSQYAQGLLSLATIKNRLAEYNESLARTRGLN 60
L AQ IMNKY+ QQ L+ + L + +QG L+ I+ + + A G
Sbjct 209 LNSDAQRIMNKYMDQNQQADLFIKAQTLANLQSQGALTEKQIQTEIQRAILASAEASGKK 268
Query 61 ISNEQSEKLSEYFIRA 76
I N + + ++ I+A
Sbjct 269 IDNRVASETADSLIKA 284
Lambda K H a alpha
0.318 0.128 0.350 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 12901056