bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-36_CDS_annotation_glimmer3.pl_2_4
Length=92
Score E
Sequences producing significant alignments: (Bits) Value
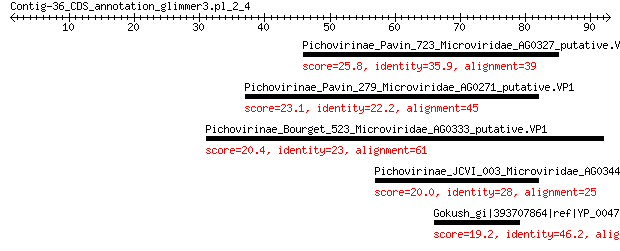
Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1 25.8 0.066
Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1 23.1 0.55
Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1 20.4 4.9
Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1 20.0 6.2
Gokush_gi|393707864|ref|YP_004732986.1|_viral_coat_protein_VP1_... 19.2 9.7
> Pichovirinae_Pavin_723_Microviridae_AG0327_putative.VP1
Length=508
Score = 25.8 bits (55), Expect = 0.066, Method: Composition-based stats.
Identities = 14/40 (35%), Positives = 20/40 (50%), Gaps = 1/40 (3%)
Query 46 NTDVAPAE-YQCVKKRKLENGYFYASVRLMHDGYAIDAEI 84
NTD E +++R E+ YF AS+ G A+D I
Sbjct 170 NTDAGDRERLTTLRQRAWEHDYFTASLPFAQKGAAVDIPI 209
> Pichovirinae_Pavin_279_Microviridae_AG0271_putative.VP1
Length=503
Score = 23.1 bits (48), Expect = 0.55, Method: Composition-based stats.
Identities = 10/45 (22%), Positives = 21/45 (47%), Gaps = 0/45 (0%)
Query 37 ISRAKETVANTDVAPAEYQCVKKRKLENGYFYASVRLMHDGYAID 81
++ +A+ + + ++KR E+ YF A + G A+D
Sbjct 154 VTAVNYNLADGNQGIGTFGTLRKRAYEHDYFTACLPFAQKGAAVD 198
> Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1
Length=518
Score = 20.4 bits (41), Expect = 4.9, Method: Composition-based stats.
Identities = 14/61 (23%), Positives = 24/61 (39%), Gaps = 5/61 (8%)
Query 31 PMLKSAISRAKETVANTDVAPAEYQCVKKRKLENGYFYASVRLMHDGYAIDAEIWKTQVH 90
P+L S + N P VKKR ++ YF + + G A+ I ++
Sbjct 168 PVLDSLTDGENDPFRNLAAGP-----VKKRAWQHDYFTSCLPWAQKGDAVTIPIGDVTIN 222
Query 91 H 91
+
Sbjct 223 Y 223
> Pichovirinae_JCVI_003_Microviridae_AG0344_putative.VP1
Length=505
Score = 20.0 bits (40), Expect = 6.2, Method: Composition-based stats.
Identities = 7/25 (28%), Positives = 14/25 (56%), Gaps = 0/25 (0%)
Query 57 VKKRKLENGYFYASVRLMHDGYAID 81
++ R E+ YF +++ G A+D
Sbjct 176 MRLRAFEHDYFTSALPFAQKGNAVD 200
> Gokush_gi|393707864|ref|YP_004732986.1|_viral_coat_protein_VP1_[Microviridae_phi-CA82]
Length=565
Score = 19.2 bits (38), Expect = 9.7, Method: Composition-based stats.
Identities = 6/13 (46%), Positives = 10/13 (77%), Gaps = 0/13 (0%)
Query 66 YFYASVRLMHDGY 78
+FYA R++ DG+
Sbjct 53 WFYAPNRILWDGW 65
Lambda K H a alpha
0.317 0.129 0.370 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 4285192