bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_5
Length=166
Score E
Sequences producing significant alignments: (Bits) Value
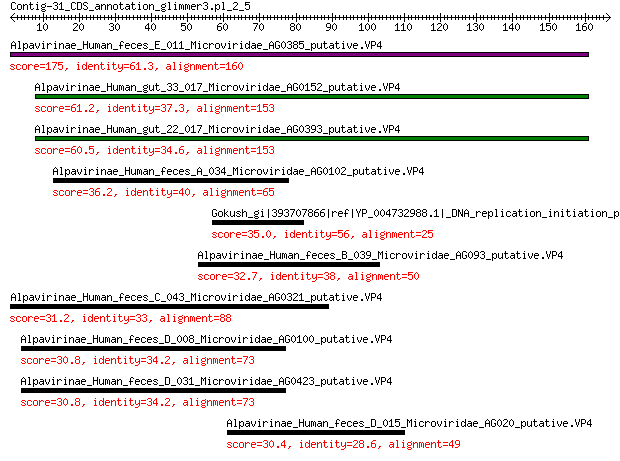
Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4 175 2e-53
Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4 61.2 4e-13
Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4 60.5 6e-13
Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4 36.2 8e-05
Gokush_gi|393707866|ref|YP_004732988.1|_DNA_replication_initiat... 35.0 2e-04
Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4 32.7 0.001
Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4 31.2 0.004
Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4 30.8 0.004
Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4 30.8 0.004
Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4 30.4 0.006
> Alpavirinae_Human_feces_E_011_Microviridae_AG0385_putative.VP4
Length=611
Score = 175 bits (444), Expect = 2e-53, Method: Compositional matrix adjust.
Identities = 98/162 (60%), Positives = 124/162 (77%), Gaps = 2/162 (1%)
Query 1 MQPKEELLTKYLHTECLRPRRVTNPYTADVLFSPcgccaacvankanvataYVQNMASYF 60
M PKEELLTKYL TECLRP+R+ NPY+ DV+F+PCG C +C+ NK+N ATAY NMA++F
Sbjct 1 MIPKEELLTKYLFTECLRPQRIVNPYSHDVIFAPCGHCKSCIMNKSNFATAYAMNMATHF 60
Query 61 KFCYFVTLTYADTFLPMVDVCAVERTGNRYL--EYADTLLPSSDPRDLKEEWYHDRDLSL 118
K+CYFVTLTY D FLP + V V R+GNRYL E +T++ +SDPR L E+YHDRD SL
Sbjct 61 KYCYFVTLTYKDIFLPYLSVEVVRRSGNRYLFDENFETMVSTSDPRFLTPEYYHDRDFSL 120
Query 119 DSNENLVRQQYEIGFKYVPRKVSVKVKNSTVYRSFDDVPFEF 160
DS +N V Q +++GF+ +PR VSVK K S +RSFDD P +F
Sbjct 121 DSAQNEVEQVFDVGFQSIPRDVSVKSKGSFRFRSFDDEPLKF 162
> Alpavirinae_Human_gut_33_017_Microviridae_AG0152_putative.VP4
Length=565
Score = 61.2 bits (147), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 57/153 (37%), Positives = 79/153 (52%), Gaps = 19/153 (12%)
Query 8 LTKYLHTECLRPRRVTNPYTADVLFSPcgccaacvankanvataYVQNMASYFKFCYFVT 67
L L T C PR V N YT + + CG C +CV ++ + T + ++ F++ YFVT
Sbjct 6 LQNKLVTRCQNPRTVVNKYTHEPVVVSCGACPSCVLRRSGIQTNLLTTYSAQFRYVYFVT 65
Query 68 LTYADTFLPMVDVCAVERTGNRYLEYADTLLPSSDPRDLKEEWYHDRDLSLDSNENLVRQ 127
LTYA FLP ++V +E + + AD SS P D+ + LD +N
Sbjct 66 LTYAPCFLPTLEVSVIETCTD---DIADV---SSVP-DIND---------LDPCDN---N 106
Query 128 QYEIGFKYVPRKVSVKVKNSTVYRSFDDVPFEF 160
+Y GF VPR SVK+KNSTV R+F D F
Sbjct 107 RYLFGFCSVPRSASVKLKNSTVERTFKDPEVRF 139
> Alpavirinae_Human_gut_22_017_Microviridae_AG0393_putative.VP4
Length=562
Score = 60.5 bits (145), Expect = 6e-13, Method: Compositional matrix adjust.
Identities = 53/154 (34%), Positives = 79/154 (51%), Gaps = 21/154 (14%)
Query 8 LTKYLHTECLRPRRVTNPYTADVLFSPcgccaacvankanvataYVQNMASYFKFCYFVT 67
L L T C PR V N YT + + CG C +C+ +++V T + ++ F++ YFVT
Sbjct 6 LQNKLVTRCQHPRTVVNKYTHEPVVVSCGHCPSCILRRSSVQTNLLTTYSAQFRYVYFVT 65
Query 68 LTYADTFLPMVDVCAVERTGNRYLEYAD-TLLPSSDPRDLKEEWYHDRDLSLDSNENLVR 126
LTYA +FLP ++V VE + + AD + +P+ D D +
Sbjct 66 LTYAPSFLPTLEVSVVETCTD---DIADVSCVPNIDELDASDP----------------- 105
Query 127 QQYEIGFKYVPRKVSVKVKNSTVYRSFDDVPFEF 160
Y GF+ VPR SVK+K+STV R+F D +F
Sbjct 106 NTYLFGFRSVPRSSSVKLKSSTVERTFKDPDVKF 139
> Alpavirinae_Human_feces_A_034_Microviridae_AG0102_putative.VP4
Length=539
Score = 36.2 bits (82), Expect = 8e-05, Method: Compositional matrix adjust.
Identities = 26/65 (40%), Positives = 37/65 (57%), Gaps = 0/65 (0%)
Query 13 HTECLRPRRVTNPYTADVLFSPcgccaacvankanvataYVQNMASYFKFCYFVTLTYAD 72
+ CL PR + N YT D ++ PCG C CV NKA A + ++C F+TLTY+
Sbjct 9 YGSCLHPRVIKNKYTGDPIYVPCGTCEFCVHNKAIKAELKCNVQLASSRYCEFITLTYST 68
Query 73 TFLPM 77
+LP+
Sbjct 69 EYLPV 73
> Gokush_gi|393707866|ref|YP_004732988.1|_DNA_replication_initiation_protein_[Microviridae_phi-CA82]
Length=353
Score = 35.0 bits (79), Expect = 2e-04, Method: Compositional matrix adjust.
Identities = 14/25 (56%), Positives = 18/25 (72%), Gaps = 0/25 (0%)
Query 57 ASYFKFCYFVTLTYADTFLPMVDVC 81
+ YFK+ YF+T+TY D LPM D C
Sbjct 97 SRYFKYQYFITITYDDDNLPMQDFC 121
> Alpavirinae_Human_feces_B_039_Microviridae_AG093_putative.VP4
Length=559
Score = 32.7 bits (73), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 19/50 (38%), Positives = 28/50 (56%), Gaps = 6/50 (12%)
Query 53 VQNMASYFKFCYFVTLTYADTFLPMVDVCAVERTGNRYLEYADTLLPSSD 102
V+ S FK+ YFVTLTY + +P+++ C V + EY D + S D
Sbjct 51 VKTAGSAFKYSYFVTLTYDNEHIPLMN-CKVLHS-----EYEDVVGISGD 94
> Alpavirinae_Human_feces_C_043_Microviridae_AG0321_putative.VP4
Length=546
Score = 31.2 bits (69), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 29/88 (33%), Positives = 44/88 (50%), Gaps = 4/88 (5%)
Query 1 MQPKEELLTKYLHTECLRPRRVTNPYTADVLFSPcgccaacvankanvataYVQNMASYF 60
M P +++ K L CL + V N YT +++ CG C AC++ A+ + V AS
Sbjct 1 MLPSDQI-HKELSKTCLHGKNVYNKYTGQMMYQSCGKCEACLSRLASARSIKVGVQASLS 59
Query 61 KFCYFVTLTYADTFLPMVDVCAVERTGN 88
K+ FVTLTY +P C + G+
Sbjct 60 KYVMFVTLTYDTYHVPK---CKIYSNGD 84
> Alpavirinae_Human_feces_D_008_Microviridae_AG0100_putative.VP4
Length=532
Score = 30.8 bits (68), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 41/73 (56%), Gaps = 1/73 (1%)
Query 4 KEELLTKYLHTECLRPRRVTNPYTADVLFSPcgccaacvankanvataYVQNMASYFKFC 63
++E + K + + C + NPYT +++ PCG C AC NK+ ++ V + +
Sbjct 3 QQEFIQK-VFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLISRHV 61
Query 64 YFVTLTYADTFLP 76
YFVTLTYA ++P
Sbjct 62 YFVTLTYAQRYIP 74
> Alpavirinae_Human_feces_D_031_Microviridae_AG0423_putative.VP4
Length=532
Score = 30.8 bits (68), Expect = 0.004, Method: Compositional matrix adjust.
Identities = 25/73 (34%), Positives = 41/73 (56%), Gaps = 1/73 (1%)
Query 4 KEELLTKYLHTECLRPRRVTNPYTADVLFSPcgccaacvankanvataYVQNMASYFKFC 63
++E + K + + C + NPYT +++ PCG C AC NK+ ++ V + +
Sbjct 3 QQEFIQK-VFSMCQHQVQTKNPYTGELIMVPCGTCPACRFNKSILSQNKVHAQSLVSRHV 61
Query 64 YFVTLTYADTFLP 76
YFVTLTYA ++P
Sbjct 62 YFVTLTYAQRYIP 74
> Alpavirinae_Human_feces_D_015_Microviridae_AG020_putative.VP4
Length=563
Score = 30.4 bits (67), Expect = 0.006, Method: Compositional matrix adjust.
Identities = 14/49 (29%), Positives = 24/49 (49%), Gaps = 7/49 (14%)
Query 61 KFCYFVTLTYADTFLPMVDVCAVERTGNRYLEYADTLLPSSDPRDLKEE 109
++CYFVTLTY ++P + + +E +Y LP P+ +
Sbjct 57 RYCYFVTLTYNTQYVPKMSLTQIE-------DYLSEWLPVRPPKSFGTQ 98
Lambda K H a alpha
0.323 0.139 0.426 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 11626264