bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_3
Length=365
Score E
Sequences producing significant alignments: (Bits) Value
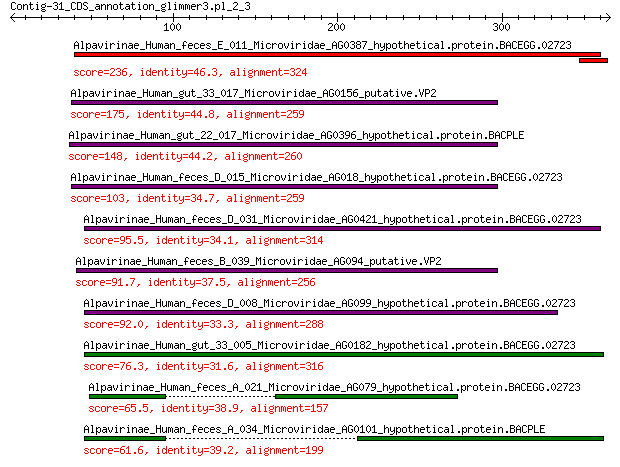
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 236 3e-75
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 175 1e-52
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 148 4e-42
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 103 1e-26
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 95.5 1e-23
Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2 91.7 2e-22
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 92.0 2e-22
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 76.3 3e-17
Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.p... 65.5 1e-13
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 61.6 2e-12
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 236 bits (603), Expect = 3e-75, Method: Compositional matrix adjust.
Identities = 148/320 (46%), Positives = 210/320 (66%), Gaps = 0/320 (0%)
Query 40 ANAMNYKINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPYlglns 99
+N+ N KIN+MNNEFNA EA+K+R Y ++MWNKTN++NSP +R+RL EAGYNPYLGL+S
Sbjct 72 SNSQNMKINRMNNEFNAREAEKARQYQSEMWNKTNDWNSPKNVRKRLQEAGYNPYLGLDS 131
Query 100 gaagsasnvgspatgsaaaaapqqSYDWSNLASSVSSAFQIANQTQQTNASVSALQGQKS 159
G+A + GS + SAA + +++S+A Q++N T+ +NA + LQGQK
Sbjct 132 SNVGTAQSAGSSSPASAAPPIQNNPIQFDGFQNALSTAIQMSNSTKVSNAEANNLQGQKG 191
Query 160 LADAETYQTLSNVDWYKLSPEYRRWLQTTGASRAALSLNTDKQQLENMQWSRNIMEAQRT 219
LADA+ TLS +DWYK +PEYR WLQTTG +RA LS NTD+Q LENM+W I AQRT
Sbjct 192 LADAQAAATLSGIDWYKFTPEYRAWLQTTGMARAQLSFNTDQQNLENMKWVNKIQRAQRT 251
Query 220 QLLLSNEQQKIMNKyidqgqqlqiqiyaaqyydlMASGHLKYQQAKTELSQRVYLAALAS 279
+LLSN+ + I+NKY+D Q LQ+++ A Q + ASG L QQ KTE+++++ A
Sbjct 252 DILLSNQAKGIINKYLDTSQSLQLKLMANQSFQAFASGRLSLQQCKTEVTKQLMNMAETE 311
Query 280 GQRISNKIAESTADSLIqaqnaqnassasYFTGFGSGSYNAGINDTWIKYNQARQGNWQY 339
G++ISNKIA TAD LI A Q ++ Y G+ + AG + + + + Y
Sbjct 312 GKKISNKIASETADQLIGALQWQYSADEMYSRGYAGYAREAGKSRGKGDVAKGQLDEYDY 371
Query 340 SKRYWDEALNAVSTVGNVVG 359
S RYW+ + +++ +GN +G
Sbjct 372 SSRYWNTGIESINRIGNGIG 391
Score = 23.1 bits (48), Expect = 4.4, Method: Compositional matrix adjust.
Identities = 10/17 (59%), Positives = 11/17 (65%), Gaps = 0/17 (0%)
Query 347 ALNAVSTVGNVVGSVRS 363
L AVS VGN+ GS S
Sbjct 56 GLGAVSGVGNIFGSALS 72
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 175 bits (444), Expect = 1e-52, Method: Compositional matrix adjust.
Identities = 116/261 (44%), Positives = 172/261 (66%), Gaps = 6/261 (2%)
Query 38 KSANAMNYKINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPYlgl 97
K N MNYKINQMNN+FN A + RD+ +MWNK N YN+ SA RQRL EAG NPYL +
Sbjct 29 KETNQMNYKINQMNNQFNERMAMQQRDFQENMWNKENTYNTASAQRQRLEEAGLNPYLMM 88
Query 98 nsgaagsasnvgspatgsaaaaapqqSY--DWSNLASSVSSAFQIANQTQQTNASVSALQ 155
N G++G + + G+ A+ S++ A Q + D+S + ++ S FQ +Q A VS +Q
Sbjct 89 NGGSSGVSQSAGTGASASSSGTAVFQPFQADFSGIQQAIGSVFQ----SQVRQAQVSQMQ 144
Query 156 GQKSLADAETYQTLSNVDWYKLSPEYRRWLQTTGASRAALSLNTDKQQLENMQWSRNIME 215
GQ++LADA+ Q LS VDW K++ E R +L+ TG +RA L + + Q+L+NM ++ +++
Sbjct 145 GQRNLADAQAMQALSQVDWSKMTKETREYLKATGLARARLGYSKEMQELDNMAFAGRLLQ 204
Query 216 AQRTQLLLSNEQQKIMNKyidqgqqlqiqiyaaqyydlMASGHLKYQQAKTELSQRVYLA 275
AQ T LL + + ++N+Y+DQ QQ + + A+ YY+ M+ GHL Y QAK ++ +
Sbjct 205 AQGTSQLLEADAKTVLNRYLDQQQQADLNVKASVYYNQMSQGHLNYNQAKKVIADEILTY 264
Query 276 ALASGQRISNKIAESTADSLI 296
A GQ++SNK+AE+TADSLI
Sbjct 265 ARIKGQKLSNKVAEATADSLI 285
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 148 bits (373), Expect = 4e-42, Method: Compositional matrix adjust.
Identities = 115/262 (44%), Positives = 168/262 (64%), Gaps = 6/262 (2%)
Query 37 TKSANAMNYKINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPYlg 96
+ N MNYKINQMNN+FN A + R++ +MWNK N YN+ SA RQRL EAG NPYL
Sbjct 28 VRETNQMNYKINQMNNQFNERMAIQQRNWQENMWNKENAYNTASAQRQRLEEAGLNPYLM 87
Query 97 lnsgaagsasnvgspatgsaaaaapqqSY--DWSNLASSVSSAFQIANQTQQTNASVSAL 154
+N G+AG A + G+ + S++ A Q + D+S + SS+ + FQ + + S L
Sbjct 88 MNGGSAGVAQSAGTGSAASSSGNAVMQPFQADYSGIGSSIGNIFQY----ELMQSEKSQL 143
Query 155 QGQKSLADAETYQTLSNVDWYKLSPEYRRWLQTTGASRAALSLNTDKQQLENMQWSRNIM 214
QG + LADA+ +TLSN+DW KL+ E R +L++TG +RA L ++Q+ +NM + ++
Sbjct 144 QGARQLADAKAMETLSNIDWGKLTDETRGFLKSTGLARAQLGYAKEQQEADNMAMTGLVL 203
Query 215 EAQRTQLLLSNEQQKIMNKyidqgqqlqiqiyaaqyydlMASGHLKYQQAKTELSQRVYL 274
AQR+ +LL NE + I+NKY+DQ QQL + + AA YY MA+G++ Y +AK L++
Sbjct 204 RAQRSGMLLDNEAKGILNKYLDQHQQLDLSVKAADYYQRMAAGYVSYAEAKKALAEEALA 263
Query 275 AALASGQRISNKIAESTADSLI 296
AA GQ ISN++A A+S I
Sbjct 264 AARTRGQNISNEVASRIAESQI 285
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 103 bits (257), Expect = 1e-26, Method: Compositional matrix adjust.
Identities = 90/260 (35%), Positives = 138/260 (53%), Gaps = 4/260 (2%)
Query 38 KSANAMNYKINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPYlgl 97
+ AN MN KINQMNNEFNA EA+K+R + DMWNK N YN+P+A R RL E GYN Y+
Sbjct 56 RKANEMNLKINQMNNEFNAKEAEKARAFQLDMWNKENAYNTPAAQRARLEEGGYNAYMNP 115
Query 98 nsgaagsasnvgspatgsaaaaapqqSYDWSNLAS-SVSSAFQIANQTQQTNASVSALQG 156
+ S + S A+ +++A D+S+L V A ++ +++ +
Sbjct 116 ADAGSASGMSSTSAASAASSAVMQGT--DFSSLGEVGVRLAQELKTFSEKKGLDIRNF-S 172
Query 157 QKSLADAETYQTLSNVDWYKLSPEYRRWLQTTGASRAALSLNTDKQQLENMQWSRNIMEA 216
K ++ + + +W +SPE R+ +G A + + ++Q N WS N++ A
Sbjct 173 LKDYLQSQIDKMKGDTNWRNVSPEAIRYNIMSGLEAAKIGMENLREQWANQVWSNNLLRA 232
Query 217 QRTQLLLSNEQQKIMNKyidqgqqlqiqiyaaqyydlMASGHLKYQQAKTELSQRVYLAA 276
LL E + I+NKY+DQ QQ + + AA Y +L+ G L +A+ LS+ V A
Sbjct 233 NVANSLLDAESKTILNKYLDQQQQADLNVKAAHYEELINRGQLHVVEARELLSREVLNYA 292
Query 277 LASGQRISNKIAESTADSLI 296
A G ISN +A +A L+
Sbjct 293 RARGLNISNWVAAKSAKGLV 312
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 95.5 bits (236), Expect = 1e-23, Method: Compositional matrix adjust.
Identities = 107/356 (30%), Positives = 177/356 (50%), Gaps = 49/356 (14%)
Query 46 KINQMNNEFNAAEAQKSRDYMT-----------DMWNKTNEYNSPSAMRQRLVEAGYNPY 94
+INQ+NNEFNA+EA K+RD+ T D WN+ N YN PSA R R+ AG+NPY
Sbjct 40 EINQINNEFNASEALKNRDFQTSEREASQQWNLDQWNRENAYNDPSAQRARMEAAGFNPY 99
Query 95 lglnsgaagsasnvgspatgsaaaaapqq---------SYDWSNLASSVSSAFQIANQTQ 145
+GS S S ++A A + D+ N+AS ++ QI N
Sbjct 100 NMNIDPGSGSTSGAQSSPGSGSSATASHTPSLPAYTGYAADFQNVASGIA---QIGNAVA 156
Query 146 Q-TNASVSALQGQKSLADAETYQTLSNVDWYKLSPEYRRWLQTTGASRAALSLNTDKQQL 204
+A +++ G + + N +W L+ Y+ Q + L ++ K++L
Sbjct 157 SGIDARLTSAYGDDLMKADIMSKIGGNSEW--LTDVYKLGRQNEAPNL--LGIDLRKKRL 212
Query 205 ENMQWSRNIME--AQRTQLLLSNEQQKIMNKyidqgqqlqiqiyaaqyydlMASGHLKYQ 262
EN+ NI AQ L L E Q+I+NK++ QQ + + A + +G L
Sbjct 213 ENLSTETNIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSEA 272
Query 263 QAKTELSQRVYLAALASGQRISNKIAESTADSLIqaqnaqnassasYFTGFGSGSYNAGI 322
Q KT++ Q+ L A A+GQ+++N++AE AD +A A+ ++A+Y+ GF + ++ AG+
Sbjct 273 QVKTQIKQQALLEAQAAGQKLNNRLAERLADYQFKAMAAEYRANAAYYNGFYNDAWQAGM 332
Query 323 NDTW-IKY--NQAR-------------QGNWQYSKRYWD--EALNA-VSTVGNVVG 359
+ ++Y N AR + +W+ + Y++ E L + +VG+VVG
Sbjct 333 SKAAQVRYESNAARIAAQMSEIFKDREKASWKNNPIYYNISELLKGLLGSVGSVVG 388
> Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2
Length=380
Score = 91.7 bits (226), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 96/271 (35%), Positives = 146/271 (54%), Gaps = 31/271 (11%)
Query 41 NAMNYKINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPYlglnsg 100
N +N ++ + N FNA +AQ RD+ MW N YNSPS+M R G NP++ ++
Sbjct 28 NKVNLRMMREQNAFNAEQAQIQRDWQKQMWGMNNAYNSPSSMISR----GLNPFVQGSAA 83
Query 101 aagsasnvgspatgsaaaaapqqSYD------WSNLAS------SVSSAFQIANQTQQTN 148
AGS S A +AA Q+Y + +LAS S +SA + ++ +QT+
Sbjct 84 MAGSKSPASGGAAATAAPVPSMQAYKPNFSSVFQSLASLAQAKASEASAGESGSRARQTD 143
Query 149 ASVSALQGQKSLADAETYQTLSNVDWYKL---SPEYRRWLQTTGASRAALSLNTDKQQLE 205
L ++ Y+ L+N W L S Y W + TG AAL +T+ Q L+
Sbjct 144 TVTPLL--------SDYYRGLTN--WKNLAIGSSGY--WNKETGRVSAALDQSTETQNLK 191
Query 206 NMQWSRNIMEAQRTQLLLSNEQQKIMNKyidqgqqlqiqiyaaqyydlMASGHLKYQQAK 265
N Q++ I AQ Q+LL+++ Q+IMNKY+DQ QQ + I A +L + G L +Q +
Sbjct 192 NAQFAERISAAQEAQILLNSDAQRIMNKYMDQNQQADLFIKAQTLANLQSQGALTEKQIQ 251
Query 266 TELSQRVYLAALASGQRISNKIAESTADSLI 296
TE+ + + +A ASG++I N++A TADSLI
Sbjct 252 TEIQRAILASAEASGKKIDNRVASETADSLI 282
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 92.0 bits (227), Expect = 2e-22, Method: Compositional matrix adjust.
Identities = 96/311 (31%), Positives = 153/311 (49%), Gaps = 34/311 (11%)
Query 46 KINQMNNEFNAAEAQKSRDYMT-----------DMWNKTNEYNSPSAMRQRLVEAGYNPY 94
+INQ+NNEFNA+EA K+RD+ T D WN+ N YN PSA R R+ AG+NPY
Sbjct 40 EINQINNEFNASEALKNRDFQTSEREASQQWNLDQWNRENAYNDPSAQRARMEAAGFNPY 99
Query 95 lglnsgaagsasnvgspatgsaaaaapqq---------SYDWSNLASSVSSAFQIANQTQ 145
+ S S S + A A + D+ N+AS ++ QI N
Sbjct 100 NMNIDAGSASTSGAQSSPGSGSQATASHTPSLPAYTGYAADFQNVASGIA---QIGNAVS 156
Query 146 Q-TNASVSALQGQKSLADAETYQTLSNVDWYKLSPEYRRWLQTTGASRAALSLNTDKQQL 204
+A +++ G + + N +W L+ Y+ Q + L ++ K++L
Sbjct 157 SGIDARLTSAYGDDLMKADIMSKIGGNSEW--LTDVYKLGRQNEAPNL--LGIDLRKKRL 212
Query 205 ENMQWSRNIME--AQRTQLLLSNEQQKIMNKyidqgqqlqiqiyaaqyydlMASGHLKYQ 262
EN+ NI AQ L L E Q+I+NK++ QQ + + A + +G L
Sbjct 213 ENLSTETNIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFAQYKAGKLSEA 272
Query 263 QAKTELSQRVYLAALASGQRISNKIAESTADSLIqaqnaqnassasYFTGFGSGSYNAGI 322
Q KT++ Q+ L A GQ+++N++AE AD +A A+ ++A+Y+ GF + ++ AG+
Sbjct 273 QVKTQIKQQALLEAQTVGQKLNNRLAERLADYQFKAMAAEYRANAAYYNGFYNDAWQAGM 332
Query 323 NDTWIKYNQAR 333
+ K QAR
Sbjct 333 S----KAAQAR 339
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 76.3 bits (186), Expect = 3e-17, Method: Compositional matrix adjust.
Identities = 100/326 (31%), Positives = 167/326 (51%), Gaps = 48/326 (15%)
Query 46 KINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPYlglnsgaagsa 105
++ Q+ NE+ ++E+QKSRD+ M++ TNE+NS R RL AG NPYL +N G+AG+A
Sbjct 35 EMQQIQNEWASSESQKSRDFAKSMFDATNEWNSAKNQRARLEAAGLNPYLMMNGGSAGTA 94
Query 106 snvgspatgsaaaaapqq-SYDWSNLASSVSSAFQIANQTQQTNASVSALQGQKSLADAE 164
S+ + A+ + Y +N+ V+S SA+ KS++DA
Sbjct 95 SSTSASTVSGASGSGGTPYQYTPTNMIGDVASY-------------ASAM---KSMSDAR 138
Query 165 TYQTLSNV-DWYKLSPEYRRWLQTTGASRAALSLNTDKQQLENMQWSRNIMEAQRTQLLL 223
T S++ D Y + P Y + T A + T +Q ++ AQ+ LLL
Sbjct 139 KTNTESDLLDKYGV-PTYESQIGKTMAD----TYFTQRQA--------DVAIAQKANLLL 185
Query 224 SNEQQKIMNKyidqgqqlqiqiyaaqyydlMASGHLKYQQAKTELSQRVYLAALASGQRI 283
N+ Q+++N Y+ + +++Q+Q+ AQY++++ G + +QAK ++ R+ + A GQ I
Sbjct 186 RNDAQEVLNMYLPEEKRIQLQMNGAQYWNMIREGVISEEQAKNLIASRLEIEARTQGQHI 245
Query 284 SNKIAESTADSLIqaqnaqnassasYFTGFGSGSYNAGIN----DTWIKYNQARQGNWQY 339
SNKIA+STADS+I A + A++ G+ S + G D W++
Sbjct 246 SNKIAKSTADSIIDATRTAKENEAAFNRGYSQFSNDVGFRTGKMDRWLQ---------DP 296
Query 340 SKRYWDEALN----AVSTVGNVVGSV 361
K WD +N + + N VGS+
Sbjct 297 VKARWDRGINNAGKFIDGLSNAVGSL 322
> Alpavirinae_Human_feces_A_021_Microviridae_AG079_hypothetical.protein.BACEGG.02723
Length=397
Score = 65.5 bits (158), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 29/46 (63%), Positives = 35/46 (76%), Gaps = 0/46 (0%)
Query 49 QMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPY 94
++N F +AEAQK+RD+ DMWNKTNEYNS + R RL EAG NPY
Sbjct 14 ELNRNFQSAEAQKNRDFEVDMWNKTNEYNSATNQRARLEEAGLNPY 59
Score = 24.3 bits (51), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 32/114 (28%), Positives = 60/114 (53%), Gaps = 6/114 (5%)
Query 162 DAETYQTLSNVDWYKLSPEYRRWLQTTGA---SRAALSLNTDKQQLENMQWSRNIMEAQR 218
D++ LS ++ +Y W Q A A S + D+Q N + +++A
Sbjct 149 DSDNSPFLSKDSASRIFGKYGNWKQAGSAVMFDNAVQSSDLDRQ---NKNLTNQMIQANM 205
Query 219 TQLLLSNEQQKIMNKyidqgqqlqiqiyaaqyydlMASGHLKYQQAKTELSQRV 272
Q+ LS++ QK++NKY+DQ + +++ I +A Y + A G L ++Q + +L Q +
Sbjct 206 IQVNLSSDAQKVINKYLDQQESVKLNIQSALYTEAAARGQLTFEQWQGQLIQNM 259
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 61.6 bits (148), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 56/156 (36%), Positives = 86/156 (55%), Gaps = 13/156 (8%)
Query 212 NIMEAQRTQLLLSNEQQKIMNKyidqgqqlqiqiyaaqyydlMASGHLKYQQAKTELSQR 271
++ AQ+ LLLS E Q++MN Y+ Q +Q+Q+ AQY++++ G +K +QAK L+ R
Sbjct 174 DVATAQKANLLLSAEAQQVMNMYLPQEKQIQLSTLGAQYWNMIRDGSIKEEQAKNLLATR 233
Query 272 VYLAALASGQRISNKIAESTADSLIqaqnaqnassasYFTGFGSGSYNAGINDTWIKYNQ 331
+ + A +GQ ISNK+A STADS+I A N + A+Y G+ S + G Y
Sbjct 234 LEIEARTAGQHISNKVARSTADSIIDATNTAKMNEAAYNRGYSQFSNDVG-------YRT 286
Query 332 ARQGNWQYS--KRYWDEALN----AVSTVGNVVGSV 361
+ W K WD +N + + N+VGSV
Sbjct 287 GKMDRWSQDPVKARWDRGINNAGKFIDGLSNIVGSV 322
Score = 50.4 bits (119), Expect = 8e-09, Method: Compositional matrix adjust.
Identities = 22/49 (45%), Positives = 36/49 (73%), Gaps = 0/49 (0%)
Query 46 KINQMNNEFNAAEAQKSRDYMTDMWNKTNEYNSPSAMRQRLVEAGYNPY 94
++ ++ NE+ ++E+QKSRD+ M++ +NE+NS + R RL EAG NPY
Sbjct 35 QMQKIQNEWASSESQKSRDFAKSMFDASNEWNSAKSQRARLEEAGLNPY 83
Lambda K H a alpha
0.311 0.121 0.348 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 31621345