bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-31_CDS_annotation_glimmer3.pl_2_2
Length=160
Score E
Sequences producing significant alignments: (Bits) Value
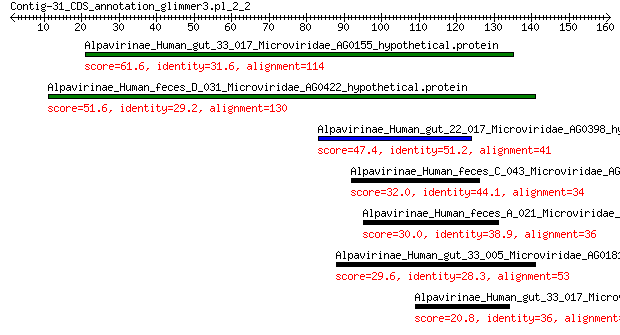
Alpavirinae_Human_gut_33_017_Microviridae_AG0155_hypothetical.p... 61.6 3e-14
Alpavirinae_Human_feces_D_031_Microviridae_AG0422_hypothetical.... 51.6 6e-11
Alpavirinae_Human_gut_22_017_Microviridae_AG0398_hypothetical.p... 47.4 7e-10
Alpavirinae_Human_feces_C_043_Microviridae_AG0323_hypothetical.... 32.0 4e-04
Alpavirinae_Human_feces_A_021_Microviridae_AG078_hypothetical.p... 30.0 0.002
Alpavirinae_Human_gut_33_005_Microviridae_AG0181_hypothetical.p... 29.6 0.008
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 20.8 7.6
> Alpavirinae_Human_gut_33_017_Microviridae_AG0155_hypothetical.protein
Length=171
Score = 61.6 bits (148), Expect = 3e-14, Method: Compositional matrix adjust.
Identities = 36/114 (32%), Positives = 68/114 (60%), Gaps = 3/114 (3%)
Query 21 DNVDLFRIEQVEHTDPLESYIRVRSDISMLFHAEATAKKIGTEGFRFLAESRRVKSSPTQ 80
D+ + R+E ++ TD +R SDI ++ H + A + G + +S++ S Q
Sbjct 44 DSTEQLRVE-IDDTDASRP-VRYTSDIRLILHNKDLASRAGVDVASKFGQSKQ-SPSQIQ 100
Query 81 AMMDKMPDDLILDTLKSRHLQQPSELLAFSEQLSALASDMEEEYEAYFKSQTVK 134
+MD M D+ +L T++SR++Q PSE+LA+S++LSA A ++E + + +++ K
Sbjct 101 QIMDTMSDEDLLATVRSRYIQSPSEILAWSKELSAYAENLESQAQELIEAENAK 154
> Alpavirinae_Human_feces_D_031_Microviridae_AG0422_hypothetical.protein
Length=150
Score = 51.6 bits (122), Expect = 6e-11, Method: Compositional matrix adjust.
Identities = 38/130 (29%), Positives = 70/130 (54%), Gaps = 10/130 (8%)
Query 11 VVEGEITLEVDNVDLFRIEQVEHTDPLESYIRVRSDISMLFHAEATAKKIGTEGFRFLAE 70
V++ I +V V++ R V+ D + IR SD+++L +AE +IG E + L
Sbjct 21 VLKSAIYCQVGPVEMLRY--VKDDDGV---IRYVSDVNLLMNAERLRNQIGEESYLNLIR 75
Query 71 SRRVKSSPTQAMMDKMPDDLILDTLKSRHLQQPSELLAFSEQLSALASDMEEEYEAYFKS 130
+ K SP +K D+ + +KSR +Q PSE+LA+ E L + + E +A +
Sbjct 76 GIQPKKSPYD---NKYTDEQLFTAIKSRFIQTPSEVLAWIESLGSAGDSIRSELDAL--T 130
Query 131 QTVKNDEESQ 140
++V+ +++S+
Sbjct 131 ESVQTNQQSE 140
> Alpavirinae_Human_gut_22_017_Microviridae_AG0398_hypothetical.protein
Length=69
Score = 47.4 bits (111), Expect = 7e-10, Method: Compositional matrix adjust.
Identities = 21/41 (51%), Positives = 32/41 (78%), Gaps = 0/41 (0%)
Query 83 MDKMPDDLILDTLKSRHLQQPSELLAFSEQLSALASDMEEE 123
MD M DD +L T++SRH+Q PSE++A+S++LSA A +E +
Sbjct 1 MDTMSDDDLLATVRSRHIQAPSEIIAWSKELSAYAEHLESQ 41
> Alpavirinae_Human_feces_C_043_Microviridae_AG0323_hypothetical.protein
Length=59
Score = 32.0 bits (71), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 15/34 (44%), Positives = 23/34 (68%), Gaps = 0/34 (0%)
Query 92 LDTLKSRHLQQPSELLAFSEQLSALASDMEEEYE 125
++T+KSR+LQ PSE+ A+ E L A + +YE
Sbjct 1 METIKSRYLQSPSEVRAWLETLVDKADVVRSDYE 34
> Alpavirinae_Human_feces_A_021_Microviridae_AG078_hypothetical.protein
Length=63
Score = 30.0 bits (66), Expect = 0.002, Method: Compositional matrix adjust.
Identities = 14/36 (39%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 95 LKSRHLQQPSELLAFSEQLSALASDMEEEYEAYFKS 130
+K R++Q +EL A+SE L+ A +++ EYE Y ++
Sbjct 4 IKPRNVQSHAELKAWSEFLTDKAKEIQTEYETYIEN 39
> Alpavirinae_Human_gut_33_005_Microviridae_AG0181_hypothetical.protein
Length=205
Score = 29.6 bits (65), Expect = 0.008, Method: Compositional matrix adjust.
Identities = 15/53 (28%), Positives = 29/53 (55%), Gaps = 0/53 (0%)
Query 88 DDLILDTLKSRHLQQPSELLAFSEQLSALASDMEEEYEAYFKSQTVKNDEESQ 140
DD +LD K R++Q +E+ ++ + L + +E + +AY S + E +Q
Sbjct 142 DDFLLDYCKDRNIQSATEMASWLDHLLSEGQSLESDLQAYADSLSASVVEPTQ 194
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 20.8 bits (42), Expect = 7.6, Method: Composition-based stats.
Identities = 9/25 (36%), Positives = 14/25 (56%), Gaps = 0/25 (0%)
Query 109 FSEQLSALASDMEEEYEAYFKSQTV 133
FS ++ SD+ AY+KS T+
Sbjct 236 FSGSSPSIVSDLPAASSAYWKSDTM 260
Lambda K H a alpha
0.314 0.129 0.344 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 10955518