bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-29_CDS_annotation_glimmer3.pl_2_2
Length=335
Score E
Sequences producing significant alignments: (Bits) Value
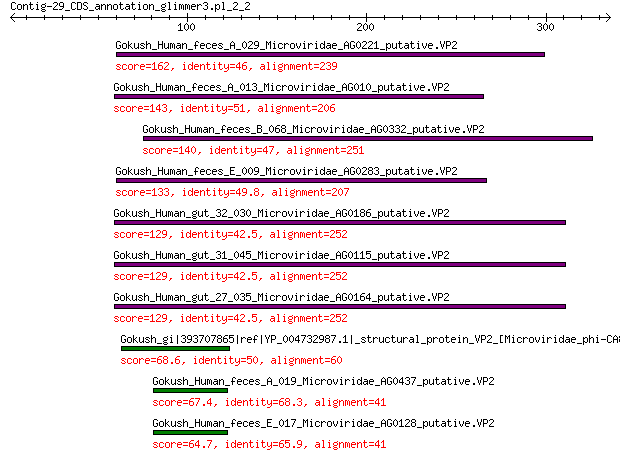
Gokush_Human_feces_A_029_Microviridae_AG0221_putative.VP2 162 2e-48
Gokush_Human_feces_A_013_Microviridae_AG010_putative.VP2 143 2e-41
Gokush_Human_feces_B_068_Microviridae_AG0332_putative.VP2 140 8e-40
Gokush_Human_feces_E_009_Microviridae_AG0283_putative.VP2 133 8e-38
Gokush_Human_gut_32_030_Microviridae_AG0186_putative.VP2 129 3e-36
Gokush_Human_gut_31_045_Microviridae_AG0115_putative.VP2 129 3e-36
Gokush_Human_gut_27_035_Microviridae_AG0164_putative.VP2 129 3e-36
Gokush_gi|393707865|ref|YP_004732987.1|_structural_protein_VP2_... 68.6 3e-15
Gokush_Human_feces_A_019_Microviridae_AG0437_putative.VP2 67.4 2e-14
Gokush_Human_feces_E_017_Microviridae_AG0128_putative.VP2 64.7 1e-13
> Gokush_Human_feces_A_029_Microviridae_AG0221_putative.VP2
Length=299
Score = 162 bits (409), Expect = 2e-48, Method: Compositional matrix adjust.
Identities = 110/239 (46%), Positives = 144/239 (60%), Gaps = 34/239 (14%)
Query 60 NNAWSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNP 119
NNAW+A+QA DKAM++N EAEK+R WQEYMSSTAHQRE+ DL AAGLNP
Sbjct 50 NNAWTAKQA-----------DKAMEFNRDEAEKSRKWQEYMSSTAHQREVKDLVAAGLNP 98
Query 120 VLSAMggngasvtsgatasssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTSAL 179
VLSAM + S + SG S DTS SS LV L GA+L+S T+L+ + A
Sbjct 99 VLSAM-----GGSGAPVTSGATASGYAPSADTSLSSGLVQLFGALLSSQTQLANKALDAQ 153
Query 180 TNLAVADKYNSVNKYLGELSSATQLKGYQISAQTALSTANISAAAQRYVSDNNLKGSLan 239
TNL+VADKY ++ + +L S QT L+TANISA A RY +D + +
Sbjct 154 TNLSVADKYTETSRAVAQLQS-----------QTQLTTANISAMASRYAADVHADAT--- 199
Query 240 aaatkiaatihaeaSKYAADKGYLSSENVANINASVNKQLKEMGIKADFDFAQMYPNNL 298
K+AA+I A A +Y D ++++ +A NA VNKQLK+M I A FD + YP++L
Sbjct 200 ----KVAASISAAAQRYGYDVMSMTNKQIAAFNADVNKQLKQMDIDAQFDLKEAYPSSL 254
> Gokush_Human_feces_A_013_Microviridae_AG010_putative.VP2
Length=276
Score = 143 bits (360), Expect = 2e-41, Method: Compositional matrix adjust.
Identities = 105/221 (48%), Positives = 145/221 (66%), Gaps = 15/221 (7%)
Query 59 ANNAWSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLN 118
ANNAWSAQQA+ R+WQ Q KAM++N++EA KNRSWQE+MS+TAHQRE+ DL AAGLN
Sbjct 23 ANNAWSAQQAQIQREWQVQQNAKAMQFNAEEAAKNRSWQEFMSNTAHQREVRDLMAAGLN 82
Query 119 PVLSAMggngasvtsgatasssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTSA 178
PVLSAM GNGA+V SGATAS SGA G TDTS S A+ NLLG++++++ L + +A
Sbjct 83 PVLSAMNGNGAAVGSGATASGVTSSGAKGDTDTSTSGAIANLLGSLVSASQALESANINA 142
Query 179 LTNLAVADKYNSVNKYLGELSSATQLKGYQISAQTALSTANISAAAQRYVSDNN------ 232
T AVADKYN++++ + E++ + L I A + A+ AAA Y +D +
Sbjct 143 RTQEAVADKYNAMSQIVAEINKSATLGSAGIHAGASKYAADRGAAATMYSADQHRAAAKY 202
Query 233 ---------LKGSLanaaatkiaatihaeaSKYAADKGYLS 264
+ GS+ +++A++ A+ A KYA+DK Y S
Sbjct 203 SADAAKLASMFGSIQSSSASRYASDQSRAAQKYASDKSYES 243
> Gokush_Human_feces_B_068_Microviridae_AG0332_putative.VP2
Length=346
Score = 140 bits (352), Expect = 8e-40, Method: Compositional matrix adjust.
Identities = 118/251 (47%), Positives = 158/251 (63%), Gaps = 24/251 (10%)
Query 75 QEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNPVLSAMggngasvtsg 134
+ AQT AM +N +EA+KNR WQ+YMS TAHQREI DL+AAGLNPVLSAMGGNGA VTSG
Sbjct 82 ESAQT--AMAFNREEAQKNRDWQQYMSDTAHQREIKDLQAAGLNPVLSAMGGNGAPVTSG 139
Query 135 atasssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTSALTNLAVADKYNSVNKY 194
ATAS A GA G TDTS S ALV+LLG+++ S T+L+ +TSA +LAVADKY + K+
Sbjct 140 ATASGYASQGAKGDTDTSASGALVSLLGSLIQSQTQLANTATSANASLAVADKYTQMQKF 199
Query 195 LGELSSATQLKGYQISAQTALSTANISAAAQRYVSDNNLKGSLanaaatkiaatihaeaS 254
+GEL + TQ L+T+ ISA A +Y +D AT+ AA IHA A
Sbjct 200 VGELQANTQ-----------LTTSKISAMASKYAADTGAS-------ATQAAAAIHAAAQ 241
Query 255 KYAADKGYLSSENVANINASVNKQLKEMGIKADFDFAQMYPNNLYQMTGATVNNLKGILG 314
KY D ++ + +A+ N+ VN L++ +FD + +P GA + L+ ++G
Sbjct 242 KYGYDVNAMTQKQIASFNSEVNWYLQQDKQAHEFDMEKYFPKT---EIGAAASGLQ-MIG 297
Query 315 DLMSQSAIDSV 325
DL++ SV
Sbjct 298 DLLTSGGSPSV 308
> Gokush_Human_feces_E_009_Microviridae_AG0283_putative.VP2
Length=294
Score = 133 bits (335), Expect = 8e-38, Method: Compositional matrix adjust.
Identities = 103/207 (50%), Positives = 131/207 (63%), Gaps = 33/207 (16%)
Query 60 NNAWSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNP 119
NNA+SA QA++LR WQE Q KAM++N+ EA KNR WQEYMS+TAHQREIADLKAAGLNP
Sbjct 46 NNAYSASQAQDLRSWQEEQNRKAMEFNAAEAAKNRDWQEYMSNTAHQREIADLKAAGLNP 105
Query 120 VLSAMggngasvtsgatasssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTSAL 179
VLSA GGNGA+VTSGATAS SGA G DTS ++AL ++LG + + L +A
Sbjct 106 VLSATGGNGAAVTSGATASGVTSSGAKGDVDTSVNAALASILGTLWNNENALKIADVNAK 165
Query 180 TNLAVADKYNSVNKYLGELSSATQLKGYQISAQTALSTANISAAAQRYVSDNNLKGSLan 239
NLAVA+KY ++N+ + QI A T+ RYVSDN+L S
Sbjct 166 NNLAVAEKYTAMNELVA-----------QIGAMTS-----------RYVSDNSLTAS--- 200
Query 240 aaatkiaatihaeaSKYAADKGYLSSE 266
+ A A++YAAD+ Y S++
Sbjct 201 --------RVMAGATQYAADRNYASTQ 219
> Gokush_Human_gut_32_030_Microviridae_AG0186_putative.VP2
Length=289
Score = 129 bits (324), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 107/256 (42%), Positives = 156/256 (61%), Gaps = 7/256 (3%)
Query 59 ANNAWSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLN 118
AN+A++A+QA+ R W E+ T + M++NS EA KNR WQE MS+TAHQRE+ DL AAGLN
Sbjct 37 ANSAFNAEQAKLQRDWTESMTARQMEFNSAEAAKNRQWQEMMSNTAHQREVRDLMAAGLN 96
Query 119 PVLSAMggngasvtsgatasssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTSA 178
PVLSAM GNGA+V SGATAS+S SG+ DT+ S A+ NLLG++L + T L + +A
Sbjct 97 PVLSAMNGNGAAVGSGATASASLGSGSKADADTAASGAIANLLGSILGAQTALQSANINA 156
Query 179 LTNLAVADKYNSVNKYLGELSSATQLKGYQISAQTALSTANISAAAQRYVSDN----NLK 234
T AVADKY ++ + ++++A +K I A A +S++A RY + ++
Sbjct 157 RTQEAVADKYTAMEHIVAQIAAAAGIKQAGIHAGATRDAAAMSSSATRYAAGQAALASMF 216
Query 235 GSLanaaatkiaatihaeaSKYAADKGYLSSENVANINASVNKQLKEMGIKADFDFAQMY 294
GS N+AAT+ +A H +KY ADK +S+ ++ N + + G K+ A M
Sbjct 217 GSSVNSAATRYSADQHLSGTKYGADKSSDASKYASDTNWDIKRTFGN-GDKS--SLAGML 273
Query 295 PNNLYQMTGATVNNLK 310
++Y + G T K
Sbjct 274 AESIYGLFGGTYRKAK 289
> Gokush_Human_gut_31_045_Microviridae_AG0115_putative.VP2
Length=289
Score = 129 bits (324), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 107/256 (42%), Positives = 156/256 (61%), Gaps = 7/256 (3%)
Query 59 ANNAWSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLN 118
AN+A++A+QA+ R W E+ T + M++NS EA KNR WQE MS+TAHQRE+ DL AAGLN
Sbjct 37 ANSAFNAEQAKLQRDWTESMTARQMEFNSAEAAKNRQWQEMMSNTAHQREVRDLMAAGLN 96
Query 119 PVLSAMggngasvtsgatasssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTSA 178
PVLSAM GNGA+V SGATAS+S SG+ DT+ S A+ NLLG++L + T L + +A
Sbjct 97 PVLSAMNGNGAAVGSGATASASLGSGSKADADTAASGAIANLLGSILGAQTALQSANINA 156
Query 179 LTNLAVADKYNSVNKYLGELSSATQLKGYQISAQTALSTANISAAAQRYVSDN----NLK 234
T AVADKY ++ + ++++A +K I A A +S++A RY + ++
Sbjct 157 RTQEAVADKYTAMEHIVAQIAAAAGIKQAGIHAGATRDAAAMSSSATRYAAGQAALASMF 216
Query 235 GSLanaaatkiaatihaeaSKYAADKGYLSSENVANINASVNKQLKEMGIKADFDFAQMY 294
GS N+AAT+ +A H +KY ADK +S+ ++ N + + G K+ A M
Sbjct 217 GSSVNSAATRYSADQHLSGTKYGADKSSDASKYASDTNWDIKRTFGN-GDKS--SLAGML 273
Query 295 PNNLYQMTGATVNNLK 310
++Y + G T K
Sbjct 274 AESIYGLFGGTYRKAK 289
> Gokush_Human_gut_27_035_Microviridae_AG0164_putative.VP2
Length=289
Score = 129 bits (324), Expect = 3e-36, Method: Compositional matrix adjust.
Identities = 107/256 (42%), Positives = 156/256 (61%), Gaps = 7/256 (3%)
Query 59 ANNAWSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLN 118
AN+A++A+QA+ R W E+ T + M++NS EA KNR WQE MS+TAHQRE+ DL AAGLN
Sbjct 37 ANSAFNAEQAKLQRDWTESMTARQMEFNSAEAAKNRQWQEMMSNTAHQREVRDLMAAGLN 96
Query 119 PVLSAMggngasvtsgatasssapsgamgstDTSGSSALVNLLGAMLTSTTELSKMSTSA 178
PVLSAM GNGA+V SGATAS+S SG+ DT+ S A+ NLLG++L + T L + +A
Sbjct 97 PVLSAMNGNGAAVGSGATASASLGSGSKADADTAASGAIANLLGSILGAQTALQSANINA 156
Query 179 LTNLAVADKYNSVNKYLGELSSATQLKGYQISAQTALSTANISAAAQRYVSDN----NLK 234
T AVADKY ++ + ++++A +K I A A +S++A RY + ++
Sbjct 157 RTQEAVADKYTAMEHIVAQIAAAAGIKQAGIHAGATRDAAAMSSSATRYAAGQAALASMF 216
Query 235 GSLanaaatkiaatihaeaSKYAADKGYLSSENVANINASVNKQLKEMGIKADFDFAQMY 294
GS N+AAT+ +A H +KY ADK +S+ ++ N + + G K+ A M
Sbjct 217 GSSVNSAATRYSADQHLSGTKYGADKSSDASKYASDTNWDIKRTFGN-GDKS--SLAGML 273
Query 295 PNNLYQMTGATVNNLK 310
++Y + G T K
Sbjct 274 AESIYGLFGGTYRKAK 289
> Gokush_gi|393707865|ref|YP_004732987.1|_structural_protein_VP2_[Microviridae_phi-CA82]
Length=234
Score = 68.6 bits (166), Expect = 3e-15, Method: Compositional matrix adjust.
Identities = 30/60 (50%), Positives = 44/60 (73%), Gaps = 0/60 (0%)
Query 63 WSAQQAEELRKWQEAQTDKAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNPVLS 122
W QA++ KW QT+K+ ++N+QEA+KNR WQE MS+TA QR++ D + AGLNP+ +
Sbjct 7 WMTAQADKQNKWNAEQTEKSNQFNAQEAQKNRDWQEQMSNTALQRKMQDAEKAGLNPIFA 66
> Gokush_Human_feces_A_019_Microviridae_AG0437_putative.VP2
Length=300
Score = 67.4 bits (163), Expect = 2e-14, Method: Compositional matrix adjust.
Identities = 28/41 (68%), Positives = 35/41 (85%), Gaps = 0/41 (0%)
Query 81 KAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNPVL 121
+AM YNS EA NR WQEYMSSTA+QR +AD++AAG+NP+L
Sbjct 132 EAMAYNSAEAAHNREWQEYMSSTAYQRAVADMRAAGINPIL 172
> Gokush_Human_feces_E_017_Microviridae_AG0128_putative.VP2
Length=300
Score = 64.7 bits (156), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 27/41 (66%), Positives = 35/41 (85%), Gaps = 0/41 (0%)
Query 81 KAMKYNSQEAEKNRSWQEYMSSTAHQREIADLKAAGLNPVL 121
+AM YNS EA NR WQE+MSSTA+QR +AD++AAG+NP+L
Sbjct 132 EAMAYNSAEAALNREWQEHMSSTAYQRAVADMRAAGINPIL 172
Lambda K H a alpha
0.306 0.119 0.324 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 28665756