bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-25_CDS_annotation_glimmer3.pl_2_3
Length=240
Score E
Sequences producing significant alignments: (Bits) Value
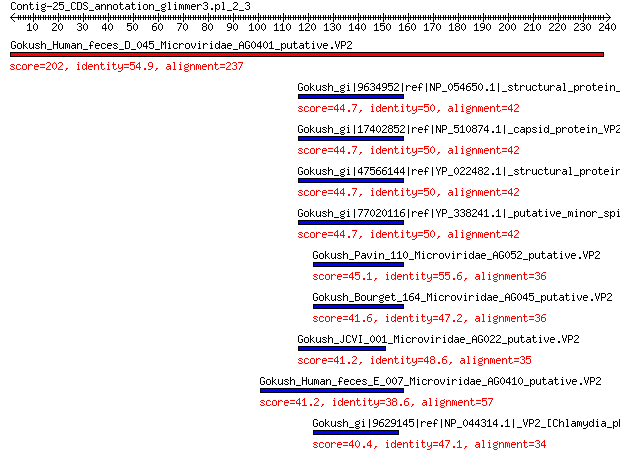
Gokush_Human_feces_D_045_Microviridae_AG0401_putative.VP2 202 9e-66
Gokush_gi|9634952|ref|NP_054650.1|_structural_protein_[Chlamydi... 44.7 1e-07
Gokush_gi|17402852|ref|NP_510874.1|_capsid_protein_VP2-related_... 44.7 1e-07
Gokush_gi|47566144|ref|YP_022482.1|_structural_protein_[Chlamyd... 44.7 1e-07
Gokush_gi|77020116|ref|YP_338241.1|_putative_minor_spike_[Chlam... 44.7 1e-07
Gokush_Pavin_110_Microviridae_AG052_putative.VP2 45.1 2e-07
Gokush_Bourget_164_Microviridae_AG045_putative.VP2 41.6 2e-06
Gokush_JCVI_001_Microviridae_AG022_putative.VP2 41.2 2e-06
Gokush_Human_feces_E_007_Microviridae_AG0410_putative.VP2 41.2 3e-06
Gokush_gi|9629145|ref|NP_044314.1|_VP2_[Chlamydia_phage_1] 40.4 5e-06
> Gokush_Human_feces_D_045_Microviridae_AG0401_putative.VP2
Length=238
Score = 202 bits (513), Expect = 9e-66, Method: Compositional matrix adjust.
Identities = 130/238 (55%), Positives = 162/238 (68%), Gaps = 5/238 (2%)
Query 1 MGFFDSLGSNSDFWQAADDIYQSSGMNQFYGQssaisakkdasiasRYPGANNPSAIINA 60
MGF+DSLGSNS+FWQ+ DDIYQSSGM+QFYGQ SAKK+A IA RYP AN+PS +I+A
Sbjct 1 MGFWDSLGSNSNFWQSVDDIYQSSGMSQFYGQDPVTSAKKNADIAVRYPDANSPSKVIDA 60
Query 61 VIGHDRDVSTSGKGTFYDEAQKEKSPAGPSFYELTRDRPSSSSQPQTYDYLDAPWASAYG 120
V+ HD+DVS SGKGT YDE QK +P+GPSFYELT DR SS ++ DYL+A A YG
Sbjct 61 VVAHDKDVSVSGKGTLYDEYQKAAAPSGPSFYELTADRHPGSS-AKSLDYLNADLAKYYG 119
Query 121 MSKETAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAGRgstagspsyissgssgssgrgs 180
M TAY EA+ NTAY+R+V DMQ AGLNPA +FGAGR S+A SY+S S GS G
Sbjct 120 MDASTAYQEALSNTAYQRAVKDMQSAGLNPAALFGAGRASSADGVSYVSQASRGSGGHSG 179
Query 181 rrgsgsgrsngKLFSGSAYSVLSALGGIVGAAATK-SAGGYWLGTSLAQGAMSALNLF 237
RR + G+ YS + G++G+A TK G++ G + A+ + L LF
Sbjct 180 RRRGSGSSAKGRFVDSGLYSAIVNGAGLIGSAITKGKPSGFFAGQAAAK---TVLGLF 234
> Gokush_gi|9634952|ref|NP_054650.1|_structural_protein_[Chlamydia_phage_2]
Length=186
Score = 44.7 bits (104), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 21/43 (49%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 116 ASAYGMSKE-TAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
A+A +++E A+ E M NTAY+R++ DM+KAGLNP + F G
Sbjct 42 ATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLNPMLAFSKG 84
> Gokush_gi|17402852|ref|NP_510874.1|_capsid_protein_VP2-related_protein_[Guinea_pig_Chlamydia_phage]
Length=186
Score = 44.7 bits (104), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 21/43 (49%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 116 ASAYGMSKE-TAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
A+A +++E A+ E M NTAY+R++ DM+KAGLNP + F G
Sbjct 42 ATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLNPMLAFSKG 84
> Gokush_gi|47566144|ref|YP_022482.1|_structural_protein_[Chlamydia_phage_3]
Length=186
Score = 44.7 bits (104), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 21/43 (49%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 116 ASAYGMSKE-TAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
A+A +++E A+ E M NTAY+R++ DM+KAGLNP + F G
Sbjct 42 ATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLNPMLAFSKG 84
> Gokush_gi|77020116|ref|YP_338241.1|_putative_minor_spike_[Chlamydia_phage_4]
Length=186
Score = 44.7 bits (104), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 21/43 (49%), Positives = 31/43 (72%), Gaps = 1/43 (2%)
Query 116 ASAYGMSKE-TAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
A+A +++E A+ E M NTAY+R++ DM+KAGLNP + F G
Sbjct 42 ATAKQIAREQMAFQERMSNTAYQRAMEDMKKAGLNPMLAFSKG 84
> Gokush_Pavin_110_Microviridae_AG052_putative.VP2
Length=257
Score = 45.1 bits (105), Expect = 2e-07, Method: Compositional matrix adjust.
Identities = 20/36 (56%), Positives = 26/36 (72%), Gaps = 0/36 (0%)
Query 122 SKETAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
S+ TA+ E M NTAY+R V+DMQ AGLNP + + G
Sbjct 34 SENTAFQERMSNTAYQRQVADMQAAGLNPMLAYIKG 69
> Gokush_Bourget_164_Microviridae_AG045_putative.VP2
Length=261
Score = 41.6 bits (96), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 17/36 (47%), Positives = 25/36 (69%), Gaps = 0/36 (0%)
Query 122 SKETAYSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
++ TA+ E M NTAY+R V D++ AGLNP + + G
Sbjct 31 NRNTAFQERMSNTAYQRQVKDLEAAGLNPMLAYVKG 66
> Gokush_JCVI_001_Microviridae_AG022_putative.VP2
Length=212
Score = 41.2 bits (95), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 17/35 (49%), Positives = 26/35 (74%), Gaps = 0/35 (0%)
Query 116 ASAYGMSKETAYSEAMENTAYRRSVSDMQKAGLNP 150
A+ Y ++ + EAM NTAY+R+++DM+ AGLNP
Sbjct 25 AADYQAQRQMDFQEAMSNTAYQRTMADMKAAGLNP 59
> Gokush_Human_feces_E_007_Microviridae_AG0410_putative.VP2
Length=270
Score = 41.2 bits (95), Expect = 3e-06, Method: Compositional matrix adjust.
Identities = 22/60 (37%), Positives = 36/60 (60%), Gaps = 3/60 (5%)
Query 101 SSSQPQTYDYLDAPWASAYGMSKETA---YSEAMENTAYRRSVSDMQKAGLNPAVIFGAG 157
S+ Q Y++++ A Y ++ A + E M +TAY+R+V DM+KAGLNP + + G
Sbjct 108 SAIQQGIYNHIENNAAMQYNSAEALANRQFQERMSSTAYQRAVEDMRKAGLNPILAYAQG 167
> Gokush_gi|9629145|ref|NP_044314.1|_VP2_[Chlamydia_phage_1]
Length=263
Score = 40.4 bits (93), Expect = 5e-06, Method: Compositional matrix adjust.
Identities = 16/34 (47%), Positives = 24/34 (71%), Gaps = 0/34 (0%)
Query 122 SKETAYSEAMENTAYRRSVSDMQKAGLNPAVIFG 155
+++ A+ E M +TA RR V D++KAGLNP + G
Sbjct 86 NRQMAFQERMSSTAVRRHVEDLKKAGLNPLLALG 119
Lambda K H a alpha
0.312 0.127 0.371 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 19051434