bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_4
Length=591
Score E
Sequences producing significant alignments: (Bits) Value
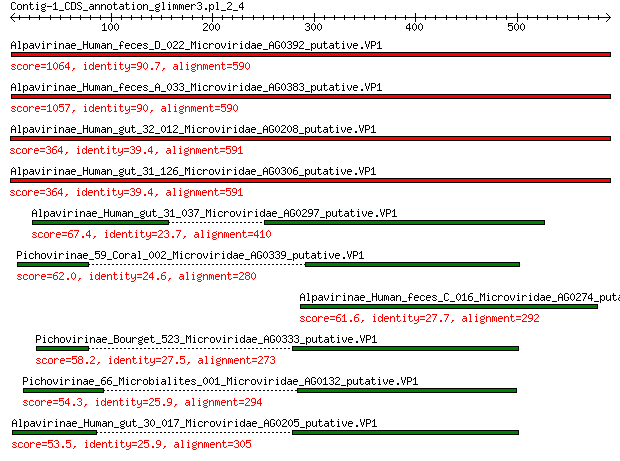
Alpavirinae_Human_feces_D_022_Microviridae_AG0392_putative.VP1 1064 0.0
Alpavirinae_Human_feces_A_033_Microviridae_AG0383_putative.VP1 1057 0.0
Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1 364 4e-119
Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1 364 4e-119
Alpavirinae_Human_gut_31_037_Microviridae_AG0297_putative.VP1 67.4 2e-13
Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1 62.0 6e-12
Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1 61.6 8e-12
Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1 58.2 1e-10
Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1 54.3 2e-09
Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1 53.5 3e-09
> Alpavirinae_Human_feces_D_022_Microviridae_AG0392_putative.VP1
Length=589
Score = 1064 bits (2751), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 535/590 (91%), Positives = 558/590 (95%), Gaps = 1/590 (0%)
Query 2 MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFM 61
MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRV+AGDDFSFQPGVGVQALPIVAPFM
Sbjct 1 MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVIAGDDFSFQPGVGVQALPIVAPFM 60
Query 62 GSVCVKKEYFFIPDRIYNIDRQLNFQGVTDTPNTVYKPSMAPPIPFDISKSMGDKVDISV 121
G+VCVKKEYFFIPDRIYNIDRQLNFQGVTDTPNTVYKPSMAPPIPFDIS G KVDISV
Sbjct 61 GNVCVKKEYFFIPDRIYNIDRQLNFQGVTDTPNTVYKPSMAPPIPFDISAPSG-KVDISV 119
Query 122 SDVQTDLPLASLGLIVGPGSLADYMGEAPGSIVTGVIDLTPyigyidtyynyylnqqysl 181
SDVQTDLPL SLG IVGPGSLADYMGEAPGSIVTGVIDLTPYIGYID YYNYYLNQQ+ L
Sbjct 120 SDVQTDLPLRSLGYIVGPGSLADYMGEAPGSIVTGVIDLTPYIGYIDIYYNYYLNQQFGL 179
Query 182 APTSLAGTASDSAMEYPYYLEVSELENYLRTIKTTPNTSPAVRRDKTASYSTNVDTALNA 241
PTSLAGT SDSAMEYPY+L+VSELE YLR IKTTPNTSPA+R D +ASYSTNV+ AL A
Sbjct 180 VPTSLAGTVSDSAMEYPYFLDVSELEGYLRAIKTTPNTSPAIREDDSASYSTNVNAALRA 239
Query 242 INSEAFMWNFFTGRQSLFQRSFPSYYLEAWLKTSSFTDAAVDVSVSGNSVSMRNITFASR 301
+ S AF+W+FFTGRQSLFQR FPSYYLEAWLKTSSF+DAAVD+S SG+SVSMRNITFASR
Sbjct 240 VASNAFLWDFFTGRQSLFQRGFPSYYLEAWLKTSSFSDAAVDISTSGSSVSMRNITFASR 299
Query 302 MQRYMDLAFAGGGRNSDFYESQFDVKLSQDNTCPAFLGSDSFDMNVNTLYQTTGFEDNSS 361
MQRYMDLAFAGGGRNSDFYE+QFDVKL+QDNTCPAFLGSDSFDMNVNTLYQTTGFEDNSS
Sbjct 300 MQRYMDLAFAGGGRNSDFYEAQFDVKLNQDNTCPAFLGSDSFDMNVNTLYQTTGFEDNSS 359
Query 362 PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVYYPSYINPTSRQISLGQQYAPA 421
PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVYYPSYINPTSRQISLGQQYAPA
Sbjct 360 PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVYYPSYINPTSRQISLGQQYAPA 419
Query 422 LDNIAMQGLKASTVFGEVQNLGANTVTYANSTLSIPGFKLQESNYVGYEPAWSELMTAVS 481
LDNIAMQGLKASTVFGEVQNL A + TYAN LSIPGFK+Q+SNYVGYEPAWSELMTAVS
Sbjct 420 LDNIAMQGLKASTVFGEVQNLSAISTTYANFILSIPGFKVQDSNYVGYEPAWSELMTAVS 479
Query 482 KPHGRLCNDLDYWVLSRDYGRNLASVMDTPAYSDFIKAAGTYVDELSLQRLTAFLKRIYV 541
KPHGRLCNDLDYWVLSRDYGRNL+ VMD+ AY DF+ AAGT +DELSLQRLTAF KRIY
Sbjct 480 KPHGRLCNDLDYWVLSRDYGRNLSYVMDSKAYGDFVSAAGTNIDELSLQRLTAFFKRIYE 539
Query 542 SPSSCPYILCGDFNYVFYDQRPTAENFVLDNVADIVVFREKSKVNVATTL 591
SPSSCPYILCGDFNYVFYDQRPTAENFVLDNVADIVVFREKSKVNVATTL
Sbjct 540 SPSSCPYILCGDFNYVFYDQRPTAENFVLDNVADIVVFREKSKVNVATTL 589
> Alpavirinae_Human_feces_A_033_Microviridae_AG0383_putative.VP1
Length=590
Score = 1057 bits (2733), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 531/590 (90%), Positives = 553/590 (94%), Gaps = 0/590 (0%)
Query 2 MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFM 61
MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFM
Sbjct 1 MFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFM 60
Query 62 GSVCVKKEYFFIPDRIYNIDRQLNFQGVTDTPNTVYKPSMAPPIPFDISKSMGDKVDISV 121
GSVCVKKEYFFIPDRIYNI+RQLNFQGVTDTPNTVYKPSMAPPIPFDIS + G++V I V
Sbjct 61 GSVCVKKEYFFIPDRIYNIERQLNFQGVTDTPNTVYKPSMAPPIPFDISVASGEEVSIPV 120
Query 122 SDVQTDLPLASLGLIVGPGSLADYMGEAPGSIVTGVIDLTPyigyidtyynyylnqqysl 181
SDVQT+LPL SLG IVGPGSLADYMGEAPGSIVTGVIDLTPYIGYID YYNYY+NQQY L
Sbjct 121 SDVQTELPLVSLGDIVGPGSLADYMGEAPGSIVTGVIDLTPYIGYIDIYYNYYVNQQYDL 180
Query 182 APTSLAGTASDSAMEYPYYLEVSELENYLRTIKTTPNTSPAVRRDKTASYSTNVDTALNA 241
PTSLAGT SDSA+EYPYYL VSELE YLRTIKTTPNTSPA+R D + SYSTNV+ AL A
Sbjct 181 VPTSLAGTKSDSAVEYPYYLSVSELETYLRTIKTTPNTSPAIRVDDSVSYSTNVEKALQA 240
Query 242 INSEAFMWNFFTGRQSLFQRSFPSYYLEAWLKTSSFTDAAVDVSVSGNSVSMRNITFASR 301
+ S AF WNFFTGRQSLFQR FPSYYLEAWLKTSSF DAAVDVS SG+SVSMRNITFASR
Sbjct 241 VGSYAFSWNFFTGRQSLFQRGFPSYYLEAWLKTSSFVDAAVDVSASGSSVSMRNITFASR 300
Query 302 MQRYMDLAFAGGGRNSDFYESQFDVKLSQDNTCPAFLGSDSFDMNVNTLYQTTGFEDNSS 361
MQRYMDLAFAGGGRNSDFYESQFDVKL QDNTCPAFLGSDSFDMNVNTLYQTTGFEDNSS
Sbjct 301 MQRYMDLAFAGGGRNSDFYESQFDVKLDQDNTCPAFLGSDSFDMNVNTLYQTTGFEDNSS 360
Query 362 PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVYYPSYINPTSRQISLGQQYAPA 421
PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVYYPSYINPTSRQISLGQQYAPA
Sbjct 361 PLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVYYPSYINPTSRQISLGQQYAPA 420
Query 422 LDNIAMQGLKASTVFGEVQNLGANTVTYANSTLSIPGFKLQESNYVGYEPAWSELMTAVS 481
LDNIAMQGLKASTVFGEVQ+LGA +YA + L IPGFKLQ+ NY+GYEPAWSELMTAVS
Sbjct 421 LDNIAMQGLKASTVFGEVQSLGATNPSYAENRLIIPGFKLQDFNYIGYEPAWSELMTAVS 480
Query 482 KPHGRLCNDLDYWVLSRDYGRNLASVMDTPAYSDFIKAAGTYVDELSLQRLTAFLKRIYV 541
KPHGRLCNDLDYWVLSRDYGRN+A VMD+ AY DFI AAG+ VDELSLQRLTAF KRIY+
Sbjct 481 KPHGRLCNDLDYWVLSRDYGRNIAHVMDSKAYKDFIAAAGSGVDELSLQRLTAFFKRIYI 540
Query 542 SPSSCPYILCGDFNYVFYDQRPTAENFVLDNVADIVVFREKSKVNVATTL 591
SPSSCPYILCGDFNYVFYDQRPTAENF+LDNVADIVVFREKSKVNVATTL
Sbjct 541 SPSSCPYILCGDFNYVFYDQRPTAENFILDNVADIVVFREKSKVNVATTL 590
> Alpavirinae_Human_gut_32_012_Microviridae_AG0208_putative.VP1
Length=604
Score = 364 bits (935), Expect = 4e-119, Method: Compositional matrix adjust.
Identities = 233/624 (37%), Positives = 333/624 (53%), Gaps = 55/624 (9%)
Query 1 MMFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPF 60
MMFL+R RNKKS L +PT+ + G L+P + TR+ AGDD F+P VQA+P+ AP
Sbjct 3 MMFLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNAPL 62
Query 61 MGSVCVKKEYFFIPDRIYNIDRQLNFQGVTDTPNTVYKPSMAPPIPFDISKSMGDKVDIS 120
+ + EYFF+PDR+YN + ++ GVTD P++V P + P + D + S
Sbjct 63 VNGFKLCLEYFFVPDRLYNWNLLMDNTGVTDNPDSVKFPQIHAPAEY-----TSDTIKFS 117
Query 121 V-SDVQTDLPLASLG-LIVGPGSLADYMGEAPGSIVT-------------------GVID 159
+ S+ A L IV PGSLADY G G T GV+D
Sbjct 118 LNSESAAKGNAARLANSIVQPGSLADYCGFPVGLFPTYEVVLDTDDRNQFCALKLLGVLD 177
Query 160 LTPyigyidtyynyylnqqyslAPTSLAGTASDSAMEYPYYLEVSELENYLRTIKTTPNT 219
+ + S PT G+ + ++Y V+ L ++L +K + +
Sbjct 178 I---FYHYYVNQQIDRFPTASFTPTLNHGSDEERNVDY----SVTMLRSFLDFVKRSADP 230
Query 220 SPAVRRDKTASYSTNVDTALNAINSEAFMWNFFTGRQSLFQRSFPSYYLEAWLKTSSFTD 279
+ A+ + T++ + N W++F R S+FQR P YYLE+WL TS + D
Sbjct 231 AGAIGQWATSNPNGNPIFG---------TWSWFCSRASIFQRCLPPYYLESWLATSGYED 281
Query 280 A--AVDVSVSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLSQDNTCPAF 337
+ VD+ G S+S RNI S +QR++DLA AGG R SD+ SQFDV + T P +
Sbjct 282 SEIKVDLGADGKSISFRNIAAQSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLY 341
Query 338 LGSDSFDMNVNTLYQTTGFEDNSSPLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVP 397
LGSD + N +YQTTG D++SPLGAF+GQ SGG FRRR+YHF ++GYF+ + S+VP
Sbjct 342 LGSDRQYLGSNVIYQTTGAGDSASPLGAFAGQSSGGETFRRRSYHFGENGYFVVMASLVP 401
Query 398 RVYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQNLGANTVTYANSTLSIP 457
V Y ++P +R+ +LG Y PALDNIAM+ L V + +L + + TLSI
Sbjct 402 DVVYSRGMDPFNREKTLGDVYVPALDNIAMEPLMIEQV-DAIPSLRSLQKVDSVYTLSIR 460
Query 458 GFKLQESNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGRNLASVMDTPAY-SDF 516
KL +++ +GY PAWS++M VS+ HGRL DL YW+LSRDYG ++ + +T +D
Sbjct 461 PDKLIKNSALGYVPAWSKVMQNVSRAHGRLTTDLKYWLLSRDYGVDVDKIKNTGVIGADV 520
Query 517 IKAAGT---------YVDELSLQRLTAFLKRIYVSPSSCPYILCGDFNYVFYDQRPTAEN 567
+K + Y+ + L RI P PY+L +N VF D A+N
Sbjct 521 LKNLASELNQAYQAGYISFEQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQN 580
Query 568 FVLDNVADIVVFREKSKVNVATTL 591
FVL + REK KVN TT+
Sbjct 581 FVLTCTFSMTCNREKGKVNTPTTI 604
> Alpavirinae_Human_gut_31_126_Microviridae_AG0306_putative.VP1
Length=604
Score = 364 bits (935), Expect = 4e-119, Method: Compositional matrix adjust.
Identities = 233/624 (37%), Positives = 333/624 (53%), Gaps = 55/624 (9%)
Query 1 MMFLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPF 60
MMFL+R RNKKS L +PT+ + G L+P + TR+ AGDD F+P VQA+P+ AP
Sbjct 3 MMFLTRSRNKKSNVNLSHVSPTTIAPGNLVPISFTRIFAGDDLRFEPSAFVQAMPMNAPL 62
Query 61 MGSVCVKKEYFFIPDRIYNIDRQLNFQGVTDTPNTVYKPSMAPPIPFDISKSMGDKVDIS 120
+ + EYFF+PDR+YN + ++ GVTD P++V P + P + D + S
Sbjct 63 VNGFKLCLEYFFVPDRLYNWNLLMDNTGVTDNPDSVKFPQIHAPAEY-----TSDTIKFS 117
Query 121 V-SDVQTDLPLASLG-LIVGPGSLADYMGEAPGSIVT-------------------GVID 159
+ S+ A L IV PGSLADY G G T GV+D
Sbjct 118 LNSESAAKGNAARLANSIVQPGSLADYCGFPVGLFPTYEVVLDTDDRNQFCALKLLGVLD 177
Query 160 LTPyigyidtyynyylnqqyslAPTSLAGTASDSAMEYPYYLEVSELENYLRTIKTTPNT 219
+ + S PT G+ + ++Y V+ L ++L +K + +
Sbjct 178 I---FYHYYVNQQIDRFPTASFTPTLNHGSDEERNVDY----SVTMLRSFLDFVKRSADP 230
Query 220 SPAVRRDKTASYSTNVDTALNAINSEAFMWNFFTGRQSLFQRSFPSYYLEAWLKTSSFTD 279
+ A+ + T++ + N W++F R S+FQR P YYLE+WL TS + D
Sbjct 231 AGAIGQWATSNPNGNPIFG---------TWSWFCSRASIFQRCLPPYYLESWLATSGYED 281
Query 280 A--AVDVSVSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLSQDNTCPAF 337
+ VD+ G S+S RNI S +QR++DLA AGG R SD+ SQFDV + T P +
Sbjct 282 SEIKVDLGADGKSISFRNIAAQSHIQRWLDLALAGGSRYSDYINSQFDVSRLKHTTSPLY 341
Query 338 LGSDSFDMNVNTLYQTTGFEDNSSPLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVP 397
LGSD + N +YQTTG D++SPLGAF+GQ SGG FRRR+YHF ++GYF+ + S+VP
Sbjct 342 LGSDRQYLGSNVIYQTTGAGDSASPLGAFAGQSSGGETFRRRSYHFGENGYFVVMASLVP 401
Query 398 RVYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQNLGANTVTYANSTLSIP 457
V Y ++P +R+ +LG Y PALDNIAM+ L V + +L + + TLSI
Sbjct 402 DVVYSRGMDPFNREKTLGDVYVPALDNIAMEPLMIEQV-DAIPSLRSLQKVDSVYTLSIR 460
Query 458 GFKLQESNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDYGRNLASVMDTPAY-SDF 516
KL +++ +GY PAWS++M VS+ HGRL DL YW+LSRDYG ++ + +T +D
Sbjct 461 PDKLIKNSALGYVPAWSKVMQNVSRAHGRLTTDLKYWLLSRDYGVDVDKIKNTGVIGADV 520
Query 517 IKAAGT---------YVDELSLQRLTAFLKRIYVSPSSCPYILCGDFNYVFYDQRPTAEN 567
+K + Y+ + L RI P PY+L +N VF D A+N
Sbjct 521 LKNLASELNQAYQAGYISFEQMDALVTLFSRIQALPEYTPYVLPNAYNDVFADTSNQAQN 580
Query 568 FVLDNVADIVVFREKSKVNVATTL 591
FVL + REK KVN TT+
Sbjct 581 FVLTCTFSMTCNREKGKVNTPTTI 604
> Alpavirinae_Human_gut_31_037_Microviridae_AG0297_putative.VP1
Length=668
Score = 67.4 bits (163), Expect = 2e-13, Method: Compositional matrix adjust.
Identities = 63/289 (22%), Positives = 122/289 (42%), Gaps = 33/289 (11%)
Query 251 FFTGRQSLFQRSFPSYYLEAWLKTSSFT-DAAVD----VSVSGNSVSMRNITFASRMQRY 305
F+ L +++ S + W+ T D ++ V+V ++ + A+++ Y
Sbjct 371 LFSPEWGLVTKTYQSDLCQNWINTEWLDGDDGINQITAVAVEDGKFTIDALNLANKVYNY 430
Query 306 MDLAFAGGGRNSDFYESQFDVKLSQDNTCPAFLGSDSFDMNVNTLYQTTGFEDNSSPLGA 365
++ G + E+ + ++ P + G S ++ + T+ D PLG+
Sbjct 431 LNRIAISDGSYRSWLETTWTGSYTERTETPIYCGGSSAEIIFQEVVSTSAATD--EPLGS 488
Query 366 FSGQLSGGTRFRRRNYHFN----DDGYFMEITSIVPRVYYPSYINPTSRQISLGQQYAPA 421
+G+ GT + + + GY + ITSI PR+ Y S+ + PA
Sbjct 489 LAGR---GTDSNHKGGYVTVRATEPGYLIGITSITPRLDYTQGNQWDVNLDSIDDLHKPA 545
Query 422 LDNIAMQGLKASTVFGEVQNLGANTVTYANSTLSIPGFKLQESNYVGYEPAWSELMTAVS 481
LD I Q L A E+ + G + N T++ ++G +PAW + MT ++
Sbjct 546 LDGIGFQDLSA-----ELLHAGTTQINTINDTIT--------QKFIGKQPAWIDYMTNIN 592
Query 482 KPHGRLCNDLDYWVLSR----DYGRNLASVMDTPAYSDFIKAAGTYVDE 526
+ +G + ++ +LSR DY +N ++ D Y D G + D+
Sbjct 593 RAYGNFRTNENFMILSRQYSLDYKKN--TIKDMTTYIDPDLYNGIFADQ 639
Score = 36.6 bits (83), Expect = 6e-04, Method: Compositional matrix adjust.
Identities = 34/139 (24%), Positives = 55/139 (40%), Gaps = 28/139 (20%)
Query 23 SASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCVKKEYFFIPDRIYNIDR 82
+ S GTL+P + D F + V P V P GS ++ + FF P R+YN
Sbjct 38 TQSVGTLVPFISIPMCKDDTFKIRLTPNVLTHPTVGPLFGSFKLQNDIFFCPFRLYN--- 94
Query 83 QLNFQGVTDTPNTVYKPSMAPPIPFDISKSMGDKVDIS-----VSDVQTDLPLASLGLIV 137
+ + I ++S+ KV ++ +DV+ D PL
Sbjct 95 -------------SWLHNNKNKIGLNMSQIKLPKVTLTAYKQYATDVKKDNPLQ------ 135
Query 138 GPGSLADYMGEAPGSIVTG 156
P SL Y+G + ++ G
Sbjct 136 -PSSLLTYLGISNVGMING 153
> Pichovirinae_59_Coral_002_Microviridae_AG0339_putative.VP1
Length=522
Score = 62.0 bits (149), Expect = 6e-12, Method: Compositional matrix adjust.
Identities = 48/211 (23%), Positives = 87/211 (41%), Gaps = 21/211 (10%)
Query 292 SMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLSQDNTC-PAFLGSDSFDMNVNTL 350
S+ ++ A R+Q +++ GG R + + F V+ S P FLG + + ++ +
Sbjct 280 SINDLRRAFRLQEWLERNARGGARYIEIIMAHFGVRSSDARLQRPEFLGGSATPITISEV 339
Query 351 YQTTGFEDNSSPLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVYYPSYINPTSR 410
QT+ +P G +G +Y + GY + I S++P+ Y I +
Sbjct 340 LQTSANNTEPTPQGNMAGHGVSVGSSNYVSYKCEEHGYIIGIMSVMPKTAYQQGIPKHFK 399
Query 411 QISLGQQYAPALDNIAMQGLKASTVFGEVQNLGANTVTYANSTLSIPGFKLQESNYVGYE 470
++ Y P+ NI Q + ++ QN +T T+ GY
Sbjct 400 KLDKFDYYWPSFANIGEQPILNEELYH--QNNATDTETF------------------GYT 439
Query 471 PAWSELMTAVSKPHGRLCNDLDYWVLSRDYG 501
P ++E S HG + LD+W + R +G
Sbjct 440 PRYAEYKYIPSTVHGEFRDSLDFWHMGRIFG 470
Score = 41.6 bits (96), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 21/70 (30%), Positives = 32/70 (46%), Gaps = 0/70 (0%)
Query 8 RNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCVK 67
R + + F L S G L+P V V GD F+ + + P++ P M V
Sbjct 11 RPQTNTFDLSHDRKFSGKIGELMPITVMEAVPGDKFNIKATNLTRFAPLITPIMHQASVY 70
Query 68 KEYFFIPDRI 77
+FF+P+RI
Sbjct 71 CHFFFVPNRI 80
> Alpavirinae_Human_feces_C_016_Microviridae_AG0274_putative.VP1
Length=641
Score = 61.6 bits (148), Expect = 8e-12, Method: Compositional matrix adjust.
Identities = 81/319 (25%), Positives = 119/319 (37%), Gaps = 69/319 (22%)
Query 287 SGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLSQDNT-CPAFLGSDSFDM 345
+G S+++ + +++Q +MD F GGR D + + + K S P FLG +
Sbjct 351 TGFSIAVPELRLKTKIQNWMDRLFISGGRVGDVFRTLWGTKSSAPYVNKPDFLGVWQASI 410
Query 346 NVNTLY-----QTTGFEDNSSPLGAFSGQLSGGTRFRRRNYHFNDDGYFMEITSIVPRVY 400
N + + +G + N L A + + +Y+ + G FM IT +VP
Sbjct 411 NPSNVRAMANGSASGEDANLGQLAACVDRYCDFSDHSGIDYYAKEPGTFMLITMLVPEPA 470
Query 401 YPSYINPTSRQISLGQQYAPALDNIAMQ-----------------GL--KASTVFGEVQN 441
Y ++P IS G + P L+ I Q GL + S FG N
Sbjct 471 YSQGLHPDLASISFGDDFNPELNGIGFQLVPRHRFSMMPRGFNLTGLDQQTSPWFG---N 527
Query 442 LGANTVTYANSTLSIPGFKLQESNYVGYEPAWSELMTAVSKPHGRLCN--DLDYWVLSRD 499
G V N+ VG E AWS L T S+ HG + YWVL+R
Sbjct 528 AGTGVVIDPNTV------------SVGEEVAWSWLRTDYSRLHGDFAQNGNYQYWVLTRR 575
Query 500 YGRNLASVMDTPAYSDFIKAAGTYVDELSLQRLTAFLKRIYVSPSSCPYILCGDFNYVFY 559
+ T Y D + GTY++ L D+ YVF
Sbjct 576 FTTYFPDA-GTGFYLD-GEYTGTYINPL-------------------------DWQYVFV 608
Query 560 DQRPTAENFVLDNVADIVV 578
DQ A NF D+ V
Sbjct 609 DQTLMAGNFSYYGTFDLKV 627
> Pichovirinae_Bourget_523_Microviridae_AG0333_putative.VP1
Length=518
Score = 58.2 bits (139), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 55/226 (24%), Positives = 96/226 (42%), Gaps = 31/226 (14%)
Query 279 DAAVDVSVSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLSQDNTC-PAF 337
D + +S + + + ++ A R+Q +++ GG R + + F VK S P +
Sbjct 268 DNSSQLSGTAEAADINSLRRAFRLQEWLERNARGGTRYIESILAHFGVKSSDARLQRPEY 327
Query 338 LGSDSFDMNVNTLYQTTGFEDNSSPLGAFSGQ---LSGGTRFRRRNYHFNDDGYFMEITS 394
LG M ++ + T + + P+G +G +SGG F+ Y+ + G+ + I S
Sbjct 328 LGGSKGKMVISEVLSTA---ETTLPVGNMAGHGISVSGGNEFK---YNVEEHGWIIGIIS 381
Query 395 IVPRVYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQNLGANTVTYANSTL 454
+ P Y I+ + ++ + P NI GE + LG YAN
Sbjct 382 VTPETAYQQGIHRSLSKLDRLDYFWPTFANI-----------GEQEVLGKEI--YAN--- 425
Query 455 SIPGFKLQESNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSRDY 500
G + E+ GY P ++E S+ G + LDYW L R +
Sbjct 426 ---GINIGET--FGYVPRYAEYKFLNSRVAGEMRTSLDYWHLGRKF 466
Score = 42.0 bits (97), Expect = 1e-05, Method: Compositional matrix adjust.
Identities = 20/51 (39%), Positives = 26/51 (51%), Gaps = 0/51 (0%)
Query 27 GTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCVKKEYFFIPDRI 77
G L PT V V GD ++ P++AP M V V YFF+P+RI
Sbjct 33 GGLYPTCVMECVPGDKVKIGTETMLRFAPLIAPVMHKVNVTTHYFFVPNRI 83
> Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1
Length=518
Score = 54.3 bits (129), Expect = 2e-09, Method: Compositional matrix adjust.
Identities = 28/79 (35%), Positives = 42/79 (53%), Gaps = 0/79 (0%)
Query 14 FKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMGSVCVKKEYFFI 73
F L +S G L+PTNV + GD F+ +P ++ LP++AP M V V YFF+
Sbjct 22 FDLSHDVKSSFKMGQLVPTNVIECMPGDLFTIRPQNMLRFLPLIAPVMHRVEVTTHYFFV 81
Query 74 PDRIYNIDRQLNFQGVTDT 92
P+R+ D + G +D
Sbjct 82 PNRLLWADWEKWITGESDV 100
Score = 42.4 bits (98), Expect = 8e-06, Method: Compositional matrix adjust.
Identities = 48/219 (22%), Positives = 82/219 (37%), Gaps = 28/219 (13%)
Query 284 VSVSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLSQDN-TCPAFLGSDS 342
V V ++V + + A R+Q +++ G R + S F V+ S P +LG
Sbjct 270 VDVQADAVDINTVRRAFRLQEWLEKNARAGTRYIESILSHFGVRSSDARLQRPEYLGGTK 329
Query 343 FDMNVNTLYQTTGFEDNSSPLGAFSGQ---LSGGTRFRRRNYHFNDDGYFMEITSIVPRV 399
+M ++ + T D + +G +G GG + Y + G+ + I ++ P
Sbjct 330 GNMVISEVLATAETTDANVAVGTMAGHGITAHGGNTIK---YRCEEHGWIIGIINVQPVT 386
Query 400 YYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQNLGANTVTYANSTLSIPGF 459
Y + + P NI Q + N YANS F
Sbjct 387 AYQQGLPREFSRFDRLDYPWPVFANIGEQEV-------------YNKELYANSPAPDEIF 433
Query 460 KLQESNYVGYEPAWSELMTAVSKPHGRLCNDLDYWVLSR 498
GY P ++E+ S+ G + + LD+W L R
Sbjct 434 --------GYIPRYAEMKFKNSRVAGEMRDTLDFWHLGR 464
> Alpavirinae_Human_gut_30_017_Microviridae_AG0205_putative.VP1
Length=647
Score = 53.5 bits (127), Expect = 3e-09, Method: Compositional matrix adjust.
Identities = 59/236 (25%), Positives = 98/236 (42%), Gaps = 35/236 (15%)
Query 279 DAAVDVSVSGNSVSMRNITFASRMQRYMDLAFAGGGRNSDFYESQFDVKLSQDNTCPAFL 338
D+ DV +G ++ + A+R+Q Y DL A G R SD+ + F K+ + P L
Sbjct 372 DSNSDVDFTGLK-TIPQLAVATRLQEYKDLIGASGSRYSDWLYTFFASKIEHVDR-PKLL 429
Query 339 GSDSFDMNVNTLYQTTGFEDNSSPLGAFSGQLSGGTRF-----RRRNYHFNDDGYFMEIT 393
S S +N + G + A GQ+ G F R + Y+F + GY ++
Sbjct 430 FSSSVMVNSQVVMNQAGQSGFAGGEAAALGQMGGSIAFNTVLGREQTYYFKEPGYIFDML 489
Query 394 SIVPRVYYPSYINPTSRQISLGQQYAPALDNIAMQGLKASTVFGEVQNLGANTVTYANST 453
+I P VY+ + I P + + P ++I Q + + LG + A S+
Sbjct 490 TIRP-VYFWTGIRPDYLEYRGPDYFNPIYNDIGYQDVP-------LWRLGFGWKSDAVSS 541
Query 454 LSIPGFKLQESNYVGYEPAWSELMTAVSKPHGRLCNDL---------DYWVLSRDY 500
+S V EP ++E ++ + G L + L YWV RD+
Sbjct 542 VS-----------VAKEPCYNEFRSSYDEVLGSLQSTLTPKASVPLQSYWVQQRDF 586
Score = 37.4 bits (85), Expect = 3e-04, Method: Compositional matrix adjust.
Identities = 20/83 (24%), Positives = 40/83 (48%), Gaps = 0/83 (0%)
Query 3 FLSRKRNKKSRFKLFSGNPTSASWGTLIPTNVTRVVAGDDFSFQPGVGVQALPIVAPFMG 62
F+S N SR+ + S + G + P + V A D + GV V++ P+ P
Sbjct 6 FISHSVNGYSRYDMPENKAFSITPGIIYPVRIQFVNARDRVTLHQGVDVRSNPLGVPSFN 65
Query 63 SVCVKKEYFFIPDRIYNIDRQLN 85
++ F++P ++Y+ + ++N
Sbjct 66 PYVLRLHRFWVPLQLYHPEMRVN 88
Lambda K H a alpha
0.318 0.133 0.394 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 54631388