bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-1_CDS_annotation_glimmer3.pl_2_3
Length=307
Score E
Sequences producing significant alignments: (Bits) Value
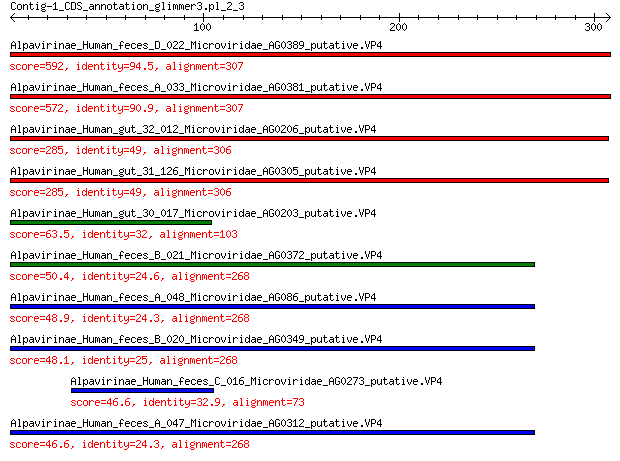
Alpavirinae_Human_feces_D_022_Microviridae_AG0389_putative.VP4 592 0.0
Alpavirinae_Human_feces_A_033_Microviridae_AG0381_putative.VP4 572 0.0
Alpavirinae_Human_gut_32_012_Microviridae_AG0206_putative.VP4 285 5e-96
Alpavirinae_Human_gut_31_126_Microviridae_AG0305_putative.VP4 285 5e-96
Alpavirinae_Human_gut_30_017_Microviridae_AG0203_putative.VP4 63.5 4e-13
Alpavirinae_Human_feces_B_021_Microviridae_AG0372_putative.VP4 50.4 6e-09
Alpavirinae_Human_feces_A_048_Microviridae_AG086_putative.VP4 48.9 2e-08
Alpavirinae_Human_feces_B_020_Microviridae_AG0349_putative.VP4 48.1 3e-08
Alpavirinae_Human_feces_C_016_Microviridae_AG0273_putative.VP4 46.6 8e-08
Alpavirinae_Human_feces_A_047_Microviridae_AG0312_putative.VP4 46.6 1e-07
> Alpavirinae_Human_feces_D_022_Microviridae_AG0389_putative.VP4
Length=307
Score = 592 bits (1526), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 290/307 (94%), Positives = 300/307 (98%), Gaps = 0/307 (0%)
Query 1 MCNKPLRITNPHYIKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCIRKRQQHWFVRAHNI 60
MCNKPLR+TNPHYIKLADQLGVE+PQFSNQ DYKLQVPCGKCVQCI+KRQQHWFVRAHNI
Sbjct 1 MCNKPLRVTNPHYIKLADQLGVEIPQFSNQPDYKLQVPCGKCVQCIKKRQQHWFVRAHNI 60
Query 61 YKRLGYNLSNSYFCTFTLKPEFYEAFCKEPYAFIRRFIDRMRKDQFLRYRNPDTGRFCYR 120
YKRLGYNLSN+YFCTFTLKPEFYEAFCKEPYAFIRRFIDRMRKDQ LRYRNPDTGRFCYR
Sbjct 61 YKRLGYNLSNTYFCTFTLKPEFYEAFCKEPYAFIRRFIDRMRKDQLLRYRNPDTGRFCYR 120
Query 121 KISFPYLFVLEVADGKRAAQrrlhsehrlhlhAIMFGCPLPWWRVRHYWMSFGLAWVSPL 180
KISFPY FVLEVADGKRAAQRRLHSEHRLHLHAIMFGCPLPWWRVRHYWMSFGLAWV+PL
Sbjct 121 KISFPYFFVLEVADGKRAAQRRLHSEHRLHLHAIMFGCPLPWWRVRHYWMSFGLAWVNPL 180
Query 181 RHFGGVRYAMKYITKKSAVHWNDVPKEILDLHGRLYVSHGFGRLSESEKDALRAYMMTGC 240
RHFG VRYAMKYITKKSAVHWNDVPKE+LDLHGRLYVSHGFGRLSESEKD LRAYMMTGC
Sbjct 181 RHFGAVRYAMKYITKKSAVHWNDVPKEVLDLHGRLYVSHGFGRLSESEKDTLRAYMMTGC 240
Query 241 KQWFSVLIDNHPYSIPRYYKLACFDKEQVRCRNDSLIPQLIWEYVLRTYPTYSNYKKQLI 300
KQWFS+LIDNHPYSIPRYYKLACF K+Q+RCRNDSLIPQL+WEYVLRTYPTYS YKKQLI
Sbjct 241 KQWFSILIDNHPYSIPRYYKLACFTKDQIRCRNDSLIPQLVWEYVLRTYPTYSYYKKQLI 300
Query 301 KQSILWQ 307
KQSILWQ
Sbjct 301 KQSILWQ 307
> Alpavirinae_Human_feces_A_033_Microviridae_AG0381_putative.VP4
Length=307
Score = 572 bits (1475), Expect = 0.0, Method: Compositional matrix adjust.
Identities = 279/307 (91%), Positives = 295/307 (96%), Gaps = 0/307 (0%)
Query 1 MCNKPLRITNPHYIKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCIRKRQQHWFVRAHNI 60
MCN+PLR+TNPHYIKLADQLGVE+PQFSNQ DYKLQVPCGKCVQCI+KRQQHWFVRAHNI
Sbjct 1 MCNRPLRVTNPHYIKLADQLGVEIPQFSNQPDYKLQVPCGKCVQCIKKRQQHWFVRAHNI 60
Query 61 YKRLGYNLSNSYFCTFTLKPEFYEAFCKEPYAFIRRFIDRMRKDQFLRYRNPDTGRFCYR 120
YKRLGYNLSN+YFCTFTLKPEFYEAFCKEPYAFIRRFIDRMRKDQ LRYR+PDTGRF YR
Sbjct 61 YKRLGYNLSNTYFCTFTLKPEFYEAFCKEPYAFIRRFIDRMRKDQSLRYRDPDTGRFRYR 120
Query 121 KISFPYLFVLEVADGKRAAQrrlhsehrlhlhAIMFGCPLPWWRVRHYWMSFGLAWVSPL 180
K+SFPYLFVLEVADGKRAAQR+L SEHRLH+HAIMFGCPLPWW VRHYWMSFGLAWVS L
Sbjct 121 KLSFPYLFVLEVADGKRAAQRKLSSEHRLHIHAIMFGCPLPWWSVRHYWMSFGLAWVSRL 180
Query 181 RHFGGVRYAMKYITKKSAVHWNDVPKEILDLHGRLYVSHGFGRLSESEKDALRAYMMTGC 240
RHFGGVRYAMKY+TKKSAVH NDVP +ILDLHGRLYVSHGFGRLSESE+DALRAYMMTGC
Sbjct 181 RHFGGVRYAMKYVTKKSAVHRNDVPNDILDLHGRLYVSHGFGRLSESERDALRAYMMTGC 240
Query 241 KQWFSVLIDNHPYSIPRYYKLACFDKEQVRCRNDSLIPQLIWEYVLRTYPTYSNYKKQLI 300
KQWFS+LIDNHPYSIPRYYKLACF K+Q+RCRNDS IPQL+WEYVLRTYPTYS YKKQLI
Sbjct 241 KQWFSILIDNHPYSIPRYYKLACFTKDQIRCRNDSFIPQLVWEYVLRTYPTYSYYKKQLI 300
Query 301 KQSILWQ 307
K SILWQ
Sbjct 301 KYSILWQ 307
> Alpavirinae_Human_gut_32_012_Microviridae_AG0206_putative.VP4
Length=306
Score = 285 bits (728), Expect = 5e-96, Method: Compositional matrix adjust.
Identities = 150/311 (48%), Positives = 203/311 (65%), Gaps = 11/311 (4%)
Query 1 MCNKPLRITNPHYIKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCIRKRQQHWFVRAHNI 60
MC + L I NPHY KLA+ V + F+ D+KL+VPCG C +C++KRQQ WF+RA ++
Sbjct 1 MCTRQLTIVNPHYKKLAEDFHVSIKCFTGLEDFKLRVPCGHCDECLKKRQQQWFLRADHV 60
Query 61 YKRLGYNLSNSYFCTFTLKPEFYEAFCKEPYAFIRRFIDRMRKDQFLRYRNPDTGRFCYR 120
KRL FCTF++KPE YE + PY IRRF+DR+RK R + +GR+ YR
Sbjct 61 VKRLKLRPDQCLFCTFSIKPEVYEQTKERPYLAIRRFMDRLRKHPRFREKGA-SGRYRYR 119
Query 121 KISFPYLFVLEVADGKRAAQrrlhsehrlhlhAIMFGCPLPWWRVRHYWMS-FGLAWVSP 179
K+ FPY+FV+E ADGKRA +R L S HR+H HAI+F CPL WW+VR W S G AWV P
Sbjct 120 KVRFPYIFVVEFADGKRARERGLESTHRMHFHAILFNCPLYWWQVRDLWESCVGRAWVDP 179
Query 180 LRHFGGVRYAMKYITKKSAVHWNDVPKEILDL----HGRLYVSHGFGRLSESEKDALRAY 235
L G+RY +KY+TK H + I D+ +G+L+VSHGFGRLS+ + +R
Sbjct 180 LHSEAGIRYVLKYMTKDCKAH-----QYISDIDARKNGKLFVSHGFGRLSKEDIKIMREN 234
Query 236 MMTGCKQWFSVLIDNHPYSIPRYYKLACFDKEQVRCRNDSLIPQLIWEYVLRTYPTYSNY 295
M+ WF V ++N YSIPRY++ ACF K++++CRNDSLIP ++W V + Y YS
Sbjct 235 MLRSSASWFCVYVNNFRYSIPRYWRTACFTKDEIKCRNDSLIPAILWSIVQQKYRGYSPV 294
Query 296 KKQLIKQSILW 306
++Q I S+LW
Sbjct 295 QQQQIYYSLLW 305
> Alpavirinae_Human_gut_31_126_Microviridae_AG0305_putative.VP4
Length=306
Score = 285 bits (728), Expect = 5e-96, Method: Compositional matrix adjust.
Identities = 150/311 (48%), Positives = 203/311 (65%), Gaps = 11/311 (4%)
Query 1 MCNKPLRITNPHYIKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCIRKRQQHWFVRAHNI 60
MC + L I NPHY KLA+ V + F+ D+KL+VPCG C +C++KRQQ WF+RA ++
Sbjct 1 MCTRQLTIVNPHYKKLAEDFHVSIKCFTGLEDFKLRVPCGHCDECLKKRQQQWFLRADHV 60
Query 61 YKRLGYNLSNSYFCTFTLKPEFYEAFCKEPYAFIRRFIDRMRKDQFLRYRNPDTGRFCYR 120
KRL FCTF++KPE YE + PY IRRF+DR+RK R + +GR+ YR
Sbjct 61 VKRLKLRPDQCLFCTFSIKPEVYEQTKERPYLAIRRFMDRLRKHPRFREKGA-SGRYRYR 119
Query 121 KISFPYLFVLEVADGKRAAQrrlhsehrlhlhAIMFGCPLPWWRVRHYWMS-FGLAWVSP 179
K+ FPY+FV+E ADGKRA +R L S HR+H HAI+F CPL WW+VR W S G AWV P
Sbjct 120 KVRFPYIFVVEFADGKRARERGLESTHRMHFHAILFNCPLYWWQVRDLWESCVGRAWVDP 179
Query 180 LRHFGGVRYAMKYITKKSAVHWNDVPKEILDL----HGRLYVSHGFGRLSESEKDALRAY 235
L G+RY +KY+TK H + I D+ +G+L+VSHGFGRLS+ + +R
Sbjct 180 LHSEAGIRYVLKYMTKDCKAH-----QYISDIDARKNGKLFVSHGFGRLSKEDIKIMREN 234
Query 236 MMTGCKQWFSVLIDNHPYSIPRYYKLACFDKEQVRCRNDSLIPQLIWEYVLRTYPTYSNY 295
M+ WF V ++N YSIPRY++ ACF K++++CRNDSLIP ++W V + Y YS
Sbjct 235 MLRSSASWFCVYVNNFRYSIPRYWRTACFTKDEIKCRNDSLIPAILWSIVQQKYRGYSPV 294
Query 296 KKQLIKQSILW 306
++Q I S+LW
Sbjct 295 QQQQIYYSLLW 305
> Alpavirinae_Human_gut_30_017_Microviridae_AG0203_putative.VP4
Length=363
Score = 63.5 bits (153), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 33/107 (31%), Positives = 60/107 (56%), Gaps = 4/107 (4%)
Query 1 MCNKPLRITNPHY----IKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCIRKRQQHWFVR 56
MC P+ I N Y I L D+ + + + + ++ VPCG C +C+R+++ W++R
Sbjct 1 MCLSPVWIRNRAYSSRTIGLTDRKVLLMNRPWDYFTQRIMVPCGHCEECLRQQRNDWYIR 60
Query 57 AHNIYKRLGYNLSNSYFCTFTLKPEFYEAFCKEPYAFIRRFIDRMRK 103
K +NS F T T+ PE+Y++ ++P +FIR + +R+R+
Sbjct 61 LERETKYHKSLHNNSVFVTITIAPEYYDSALQDPSSFIRMWFERIRR 107
> Alpavirinae_Human_feces_B_021_Microviridae_AG0372_putative.VP4
Length=332
Score = 50.4 bits (119), Expect = 6e-09, Method: Compositional matrix adjust.
Identities = 66/277 (24%), Positives = 107/277 (39%), Gaps = 48/277 (17%)
Query 1 MCNKPLRITNPHYIKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCIRKRQQHWFVRAHNI 60
MC P I N Y+ P+ ++ + CGKC++C +++Q+ W VR
Sbjct 1 MCLYPKLIRNKKYLPTKKN-NYNPPKMADPRTAYITAACGKCLECRKQKQREWLVRMSEE 59
Query 61 YKRLGYNLSNSYFCTFTLKPEFYEAF---CKE------PYAFIRRFIDRMRKDQFLRYRN 111
+ N+YF T T+ E YE CK IR ++R+RK
Sbjct 60 LR----TEPNAYFMTLTISDENYEILKNICKSEDENTIATKAIRLMLERIRK-------- 107
Query 112 PDTGRFCYRKISFPYLFVLEVADGKRAAQrrlhsehrlhlhAIMFGCPLPWWRVRHYWMS 171
G+ S + F+ E+ K RLHLH I++G +R W +
Sbjct 108 -KIGK------SIRHWFITELGHEK---------TERLHLHGIVWGIGTDQL-IREKW-N 149
Query 172 FGLAWVSPLRHFGGVRYAMKYITKKSAVHWNDVPKEILDLHGRLYVSHGFGRLSESEKDA 231
+G+ + + + Y KY+TK H N V G++ S G G DA
Sbjct 150 YGITYTGNFVNEKTINYITKYMTKIDEEHPNFV--------GKVLCSKGIGAGYTKRPDA 201
Query 232 LRAYMMTGCKQWFSVLIDNHPYSIPRYYKLACFDKEQ 268
+ G L + ++P YY+ F +++
Sbjct 202 AKHKYKKGETIETYRLRNGAKINLPIYYRNKLFTEKE 238
> Alpavirinae_Human_feces_A_048_Microviridae_AG086_putative.VP4
Length=332
Score = 48.9 bits (115), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 65/277 (23%), Positives = 105/277 (38%), Gaps = 48/277 (17%)
Query 1 MCNKPLRITNPHYIKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCIRKRQQHWFVRAHNI 60
MC P I N Y+ P+ + + CGKC++C +++Q+ W VR
Sbjct 1 MCLYPKLIRNKKYLPTKKN-NYNPPKMVDPRTAYITAACGKCLECRKQKQREWLVRMSEE 59
Query 61 YKRLGYNLSNSYFCTFTLKPEFYEAF---CKE------PYAFIRRFIDRMRKDQFLRYRN 111
+ N+YF T T+ E YE CK IR ++R+RK
Sbjct 60 LR----TEPNAYFMTLTISDENYEILKNICKSEDDNTIATKAIRLMLERIRK-------- 107
Query 112 PDTGRFCYRKISFPYLFVLEVADGKRAAQrrlhsehrlhlhAIMFGCPLPWWRVRHYWMS 171
TG+ S + F+ E+ K RLHLH I++G + W +
Sbjct 108 -KTGK------SIKHWFITELGHEK---------TERLHLHGIVWGIGTDQL-IEEKW-N 149
Query 172 FGLAWVSPLRHFGGVRYAMKYITKKSAVHWNDVPKEILDLHGRLYVSHGFGRLSESEKDA 231
+G+ + + + Y KY+TK H D G++ S G G DA
Sbjct 150 YGITYTGNYVNEKTINYVTKYMTKIDEKH--------PDFVGKVLCSRGIGAGYTKRPDA 201
Query 232 LRAYMMTGCKQWFSVLIDNHPYSIPRYYKLACFDKEQ 268
+ G L + ++P YY+ F +++
Sbjct 202 AKHKYKKGETIETYRLRNGAKINLPIYYRNKLFTEKE 238
> Alpavirinae_Human_feces_B_020_Microviridae_AG0349_putative.VP4
Length=332
Score = 48.1 bits (113), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 67/277 (24%), Positives = 108/277 (39%), Gaps = 48/277 (17%)
Query 1 MCNKPLRITNPHYIKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCIRKRQQHWFVRAHNI 60
MC P I N Y+ GV P ++ + CGKC++C +++Q+ W VR +
Sbjct 1 MCLYPKLIRNKRYLPNKKNGGVP-PVCPDERLLYVTAACGKCMECRQQKQRQWLVR---M 56
Query 61 YKRLGYNLSNSYFCTFTLKPEFYEAFC---------KEPYAFIRRFIDRMRKDQFLRYRN 111
+ L N N+YF T T+ E Y + +R ++R+RK
Sbjct 57 SEELRQN-PNAYFMTLTIDDENYNKLANICNSKDNNEIATKAVRLMLERIRK-------- 107
Query 112 PDTGRFCYRKISFPYLFVLEVADGKRAAQrrlhsehrlhlhAIMFGCPLPWWRVRHYWMS 171
TG+ S + F+ E+ K RLHLH I++G + W +
Sbjct 108 -KTGK------SIKHWFITELGHEK---------TERLHLHGIVWGIGTDQL-ISEKW-N 149
Query 172 FGLAWVSPLRHFGGVRYAMKYITKKSAVHWNDVPKEILDLHGRLYVSHGFGRLSESEKDA 231
+G + + + Y KY+TK H P D G++ S G G +DA
Sbjct 150 YGFVYTGNFVNEATINYITKYMTKVDIDH----P----DFVGQVLCSKGIGAGYTKREDA 201
Query 232 LRAYMMTGCKQWFSVLIDNHPYSIPRYYKLACFDKEQ 268
G L + ++P YY+ F +E+
Sbjct 202 NNHKYTKGKTNETYRLRNGAKINLPIYYRNQLFSEEE 238
> Alpavirinae_Human_feces_C_016_Microviridae_AG0273_putative.VP4
Length=304
Score = 46.6 bits (109), Expect = 8e-08, Method: Compositional matrix adjust.
Identities = 24/73 (33%), Positives = 36/73 (49%), Gaps = 2/73 (3%)
Query 32 DYKLQVPCGKCVQCIRKRQQHWFVRAHNIYKRLGYNLSNSYFCTFTLKPEFYEAFCKEPY 91
DY L+VPCG C C + + +R +Y+ Y F T T + + F K+
Sbjct 21 DYNLEVPCGYCHSCQKSYNNQYRIRL--LYELRKYPSGTCLFVTLTFDDDNLKKFSKDTN 78
Query 92 AFIRRFIDRMRKD 104
+R F+DR+RKD
Sbjct 79 KAVRLFLDRLRKD 91
> Alpavirinae_Human_feces_A_047_Microviridae_AG0312_putative.VP4
Length=332
Score = 46.6 bits (109), Expect = 1e-07, Method: Compositional matrix adjust.
Identities = 65/277 (23%), Positives = 107/277 (39%), Gaps = 48/277 (17%)
Query 1 MCNKPLRITNPHYIKLADQLGVEVPQFSNQSDYKLQVPCGKCVQCIRKRQQHWFVRAHNI 60
MC P I N Y+ GV PQ ++ + CGKC++C +++Q+ W VR
Sbjct 1 MCLYPKLIKNRRYVPNKKNGGVP-PQCPDERLRYITAACGKCMECRQQKQRQWLVRMSEE 59
Query 61 YKRLGYNLSNSYFCTFTLKPEFYEAFCKE---------PYAFIRRFIDRMRKDQFLRYRN 111
++ N+YF T T+ + Y IR ++R+RK
Sbjct 60 LRQ----EPNAYFITLTIDDKSYSELSNTYNITDNNEIATKAIRLCLERIRK-------- 107
Query 112 PDTGRFCYRKISFPYLFVLEVADGKRAAQrrlhsehrlhlhAIMFGCPLPWWRVRHYWMS 171
TG+ S + F+ E+ K RLHLH I++G + W +
Sbjct 108 -QTGK------SIKHWFITELGHEK---------TERLHLHGIVWGIGTDKL-ITSKW-N 149
Query 172 FGLAWVSPLRHFGGVRYAMKYITKKSAVHWNDVPKEILDLHGRLYVSHGFGRLSESEKDA 231
+G+ + + ++Y KY+TK H D G++ S G G DA
Sbjct 150 YGITFTGFFVNEKTIQYITKYMTKIDEQH--------KDFIGKVLCSKGIGAGYIKRDDA 201
Query 232 LRAYMMTGCKQWFSVLIDNHPYSIPRYYKLACFDKEQ 268
+ G L + ++P YY+ F +E+
Sbjct 202 KKHTYKRGETIETYRLRNGSKINLPIYYRNQLFTEEE 238
Lambda K H a alpha
0.328 0.141 0.480 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 25893499