bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-18_CDS_annotation_glimmer3.pl_2_5
Length=361
Score E
Sequences producing significant alignments: (Bits) Value
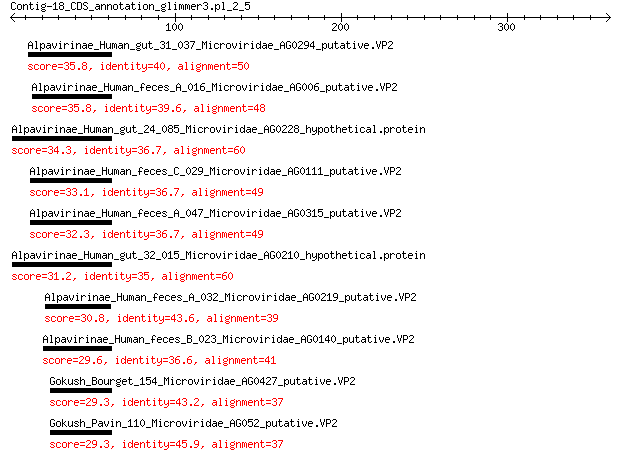
Alpavirinae_Human_gut_31_037_Microviridae_AG0294_putative.VP2 35.8 4e-04
Alpavirinae_Human_feces_A_016_Microviridae_AG006_putative.VP2 35.8 4e-04
Alpavirinae_Human_gut_24_085_Microviridae_AG0228_hypothetical.p... 34.3 0.001
Alpavirinae_Human_feces_C_029_Microviridae_AG0111_putative.VP2 33.1 0.003
Alpavirinae_Human_feces_A_047_Microviridae_AG0315_putative.VP2 32.3 0.005
Alpavirinae_Human_gut_32_015_Microviridae_AG0210_hypothetical.p... 31.2 0.010
Alpavirinae_Human_feces_A_032_Microviridae_AG0219_putative.VP2 30.8 0.013
Alpavirinae_Human_feces_B_023_Microviridae_AG0140_putative.VP2 29.6 0.031
Gokush_Bourget_154_Microviridae_AG0427_putative.VP2 29.3 0.043
Gokush_Pavin_110_Microviridae_AG052_putative.VP2 29.3 0.044
> Alpavirinae_Human_gut_31_037_Microviridae_AG0294_putative.VP2
Length=360
Score = 35.8 bits (81), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 20/50 (40%), Positives = 29/50 (58%), Gaps = 4/50 (8%)
Query 12 AQINYKYGEMAAENAFERQQVLYNRTYQDQSYANQVAQMDAAGLSPGLMY 61
A IN +Y A+ + + ++N T +Y NQVA M AAGL+P L+Y
Sbjct 41 AMINNQYALGMAKESHNMNKDMWNYT----NYENQVAHMKAAGLNPALLY 86
> Alpavirinae_Human_feces_A_016_Microviridae_AG006_putative.VP2
Length=367
Score = 35.8 bits (81), Expect = 4e-04, Method: Compositional matrix adjust.
Identities = 19/48 (40%), Positives = 28/48 (58%), Gaps = 4/48 (8%)
Query 14 INYKYGEMAAENAFERQQVLYNRTYQDQSYANQVAQMDAAGLSPGLMY 61
+ Y+YG+ AA A R ++N+T ++ Q M+ AGLS GLMY
Sbjct 72 MQYQYGQAAANEAQRRNMEMWNQT----NFGAQRQHMEDAGLSVGLMY 115
> Alpavirinae_Human_gut_24_085_Microviridae_AG0228_hypothetical.protein
Length=335
Score = 34.3 bits (77), Expect = 0.001, Method: Compositional matrix adjust.
Identities = 22/60 (37%), Positives = 32/60 (53%), Gaps = 8/60 (13%)
Query 2 QQQKEMMENAAQINYKYGEMAAENAFERQQVLYNRTYQDQSYANQVAQMDAAGLSPGLMY 61
+ +KEMM + Y+Y E AA N R ++ +T Y QV Q++ AGL+ LMY
Sbjct 53 EHEKEMM----GLQYQYNEAAANNNMTRALEMWRKT----GYEAQVQQIENAGLNEALMY 104
> Alpavirinae_Human_feces_C_029_Microviridae_AG0111_putative.VP2
Length=329
Score = 33.1 bits (74), Expect = 0.003, Method: Compositional matrix adjust.
Identities = 18/49 (37%), Positives = 29/49 (59%), Gaps = 4/49 (8%)
Query 13 QINYKYGEMAAENAFERQQVLYNRTYQDQSYANQVAQMDAAGLSPGLMY 61
+I +Y E A+N +R + L++ T +Y NQ ++ AGL+P LMY
Sbjct 18 EIQNRYNEQMAKNNQQRNKDLWDYT----NYENQKQHIENAGLNPALMY 62
> Alpavirinae_Human_feces_A_047_Microviridae_AG0315_putative.VP2
Length=325
Score = 32.3 bits (72), Expect = 0.005, Method: Compositional matrix adjust.
Identities = 18/49 (37%), Positives = 28/49 (57%), Gaps = 4/49 (8%)
Query 13 QINYKYGEMAAENAFERQQVLYNRTYQDQSYANQVAQMDAAGLSPGLMY 61
+I +Y E A+N +R + L++ T +Y NQ + AGL+P LMY
Sbjct 2 EIQNRYNEQMAKNNQQRNKDLWDYT----NYENQKQHIKNAGLNPALMY 46
> Alpavirinae_Human_gut_32_015_Microviridae_AG0210_hypothetical.protein
Length=341
Score = 31.2 bits (69), Expect = 0.010, Method: Compositional matrix adjust.
Identities = 21/60 (35%), Positives = 31/60 (52%), Gaps = 8/60 (13%)
Query 2 QQQKEMMENAAQINYKYGEMAAENAFERQQVLYNRTYQDQSYANQVAQMDAAGLSPGLMY 61
+ +KEMM + Y+Y E AA N R ++ +T Y Q Q++ AGL+ LMY
Sbjct 53 EHEKEMM----GLQYQYNEAAANNNMTRALEMWEKT----GYEAQGQQIENAGLNKALMY 104
> Alpavirinae_Human_feces_A_032_Microviridae_AG0219_putative.VP2
Length=355
Score = 30.8 bits (68), Expect = 0.013, Method: Compositional matrix adjust.
Identities = 17/39 (44%), Positives = 24/39 (62%), Gaps = 4/39 (10%)
Query 22 AAENAFERQQVLYNRTYQDQSYANQVAQMDAAGLSPGLM 60
AAE + E + +++ T Y NQV QM AAGL+P L+
Sbjct 71 AAEQSQEYAKEMFDYT----GYENQVKQMKAAGLNPALL 105
> Alpavirinae_Human_feces_B_023_Microviridae_AG0140_putative.VP2
Length=295
Score = 29.6 bits (65), Expect = 0.031, Method: Compositional matrix adjust.
Identities = 15/46 (33%), Positives = 26/46 (57%), Gaps = 5/46 (11%)
Query 21 MAAENAFERQQVLYNR-----TYQDQSYANQVAQMDAAGLSPGLMY 61
MA + + ++Q Y++ + +Y +QV M AAGL+P L+Y
Sbjct 19 MALQAKYNKEQADYSQQLALDMWNATNYESQVEHMKAAGLNPALLY 64
> Gokush_Bourget_154_Microviridae_AG0427_putative.VP2
Length=253
Score = 29.3 bits (64), Expect = 0.043, Method: Compositional matrix adjust.
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 4/41 (10%)
Query 25 NAFERQQVLYNRTYQDQ----SYANQVAQMDAAGLSPGLMY 61
NA RQ N +Q++ +Y QV ++AAGL+P L Y
Sbjct 28 NATNRQLAAENSAFQERLSNTAYQRQVKDLEAAGLNPMLAY 68
> Gokush_Pavin_110_Microviridae_AG052_putative.VP2
Length=257
Score = 29.3 bits (64), Expect = 0.044, Method: Compositional matrix adjust.
Identities = 17/41 (41%), Positives = 22/41 (54%), Gaps = 4/41 (10%)
Query 25 NAFERQQVLYNRTYQDQ----SYANQVAQMDAAGLSPGLMY 61
N RQ N +Q++ +Y QVA M AAGL+P L Y
Sbjct 26 NTTNRQIASENTAFQERMSNTAYQRQVADMQAAGLNPMLAY 66
Lambda K H a alpha
0.312 0.127 0.356 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 31192581