bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-16_CDS_annotation_glimmer3.pl_2_4
Length=65
Score E
Sequences producing significant alignments: (Bits) Value
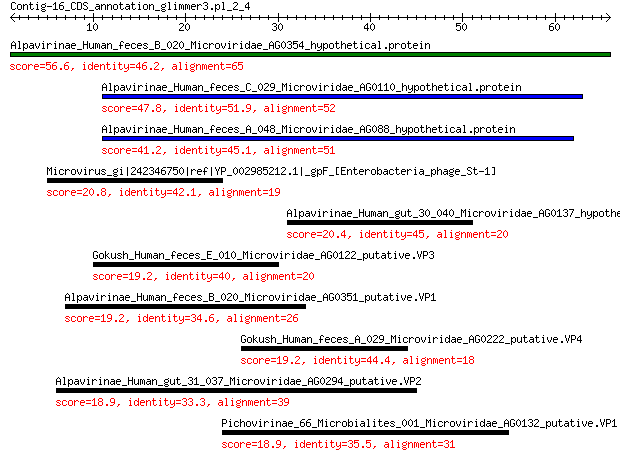
Alpavirinae_Human_feces_B_020_Microviridae_AG0354_hypothetical.... 56.6 8e-14
Alpavirinae_Human_feces_C_029_Microviridae_AG0110_hypothetical.... 47.8 9e-11
Alpavirinae_Human_feces_A_048_Microviridae_AG088_hypothetical.p... 41.2 2e-08
Microvirus_gi|242346750|ref|YP_002985212.1|_gpF_[Enterobacteria... 20.8 1.7
Alpavirinae_Human_gut_30_040_Microviridae_AG0137_hypothetical.p... 20.4 1.8
Gokush_Human_feces_E_010_Microviridae_AG0122_putative.VP3 19.2 4.5
Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1 19.2 5.2
Gokush_Human_feces_A_029_Microviridae_AG0222_putative.VP4 19.2 5.3
Alpavirinae_Human_gut_31_037_Microviridae_AG0294_putative.VP2 18.9 7.0
Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1 18.9 7.1
> Alpavirinae_Human_feces_B_020_Microviridae_AG0354_hypothetical.protein
Length=80
Score = 56.6 bits (135), Expect = 8e-14, Method: Compositional matrix adjust.
Identities = 30/65 (46%), Positives = 43/65 (66%), Gaps = 0/65 (0%)
Query 1 MKTFVWYNSHTNEETEGFEIIDDEIQKHYIFTKKERRLLCEKIYDKNEDYEKWMKSLQLN 60
K ++W+N HT E EGF+I +D I K Y K ERRLL E YD + +++ ++LQLN
Sbjct 13 FKRWLWHNLHTGETLEGFQIWEDTIFKSYNIKKTERRLLVEITYDLEKSQKEYFENLQLN 72
Query 61 LFNDE 65
LF++E
Sbjct 73 LFDNE 77
> Alpavirinae_Human_feces_C_029_Microviridae_AG0110_hypothetical.protein
Length=54
Score = 47.8 bits (112), Expect = 9e-11, Method: Compositional matrix adjust.
Identities = 27/53 (51%), Positives = 35/53 (66%), Gaps = 2/53 (4%)
Query 11 TNEETEGFEIIDDEIQKHYIFTKKERRLLCEKIYDKN-EDYEKWMKSLQLNLF 62
E T+GFEI +D + K Y +KER+L+CEK YDK+ ED +KW QL LF
Sbjct 3 PGEVTQGFEIFEDTVIKEYNLNRKERKLICEKTYDKDLEDLKKWNHK-QLKLF 54
> Alpavirinae_Human_feces_A_048_Microviridae_AG088_hypothetical.protein
Length=54
Score = 41.2 bits (95), Expect = 2e-08, Method: Compositional matrix adjust.
Identities = 23/51 (45%), Positives = 31/51 (61%), Gaps = 0/51 (0%)
Query 11 TNEETEGFEIIDDEIQKHYIFTKKERRLLCEKIYDKNEDYEKWMKSLQLNL 61
E T GFEI +D I K Y +KERRL+ EK YD+ +D + + + QL L
Sbjct 3 PGECTNGFEIFEDTIIKEYNLNRKERRLIIEKTYDEQKDLLRHVLNQQLKL 53
> Microvirus_gi|242346750|ref|YP_002985212.1|_gpF_[Enterobacteria_phage_St-1]
Length=431
Score = 20.8 bits (42), Expect = 1.7, Method: Composition-based stats.
Identities = 8/25 (32%), Positives = 13/25 (52%), Gaps = 6/25 (24%)
Query 5 VWYNSHTN------EETEGFEIIDD 23
+WY +H + + EGF +DD
Sbjct 350 IWYRTHPDYVNYKYQLLEGFPFLDD 374
> Alpavirinae_Human_gut_30_040_Microviridae_AG0137_hypothetical.protein
Length=102
Score = 20.4 bits (41), Expect = 1.8, Method: Compositional matrix adjust.
Identities = 9/20 (45%), Positives = 14/20 (70%), Gaps = 0/20 (0%)
Query 31 FTKKERRLLCEKIYDKNEDY 50
F+KKE +L +KI +N +Y
Sbjct 75 FSKKETVILGQKIKKENPNY 94
> Gokush_Human_feces_E_010_Microviridae_AG0122_putative.VP3
Length=149
Score = 19.2 bits (38), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 8/21 (38%), Positives = 11/21 (52%), Gaps = 1/21 (5%)
Query 10 HTNEETEGFEIIDDEI-QKHY 29
HTN EG D + Q+H+
Sbjct 7 HTNATAEGIVFTDPSMTQQHF 27
> Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1
Length=651
Score = 19.2 bits (38), Expect = 5.2, Method: Compositional matrix adjust.
Identities = 9/26 (35%), Positives = 12/26 (46%), Gaps = 0/26 (0%)
Query 7 YNSHTNEETEGFEIIDDEIQKHYIFT 32
Y+S TN D I+ +YIF
Sbjct 595 YDSGTNPRIADLTTYIDPIKYNYIFA 620
> Gokush_Human_feces_A_029_Microviridae_AG0222_putative.VP4
Length=343
Score = 19.2 bits (38), Expect = 5.3, Method: Composition-based stats.
Identities = 8/18 (44%), Positives = 11/18 (61%), Gaps = 0/18 (0%)
Query 26 QKHYIFTKKERRLLCEKI 43
QKH + +RR L E+I
Sbjct 314 QKHSNLDEYDRRALAERI 331
> Alpavirinae_Human_gut_31_037_Microviridae_AG0294_putative.VP2
Length=360
Score = 18.9 bits (37), Expect = 7.0, Method: Composition-based stats.
Identities = 13/42 (31%), Positives = 20/42 (48%), Gaps = 3/42 (7%)
Query 6 WYNSHTNE---ETEGFEIIDDEIQKHYIFTKKERRLLCEKIY 44
WYN+ TN T + ++ + KER+LL + IY
Sbjct 262 WYNAGTNRMNATTAADHVANELFKTMGELDIKERQLLKDWIY 303
> Pichovirinae_66_Microbialites_001_Microviridae_AG0132_putative.VP1
Length=518
Score = 18.9 bits (37), Expect = 7.1, Method: Composition-based stats.
Identities = 11/31 (35%), Positives = 14/31 (45%), Gaps = 9/31 (29%)
Query 24 EIQKHYIFTKKERRLLCEKIYDKNEDYEKWM 54
E+ HY F RLL D+EKW+
Sbjct 73 EVTTHYFFVPN--RLLWA-------DWEKWI 94
Lambda K H a alpha
0.317 0.135 0.410 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 3675489