bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
Query= Contig-15_CDS_annotation_glimmer3.pl_2_6
Length=211
Score E
Sequences producing significant alignments: (Bits) Value
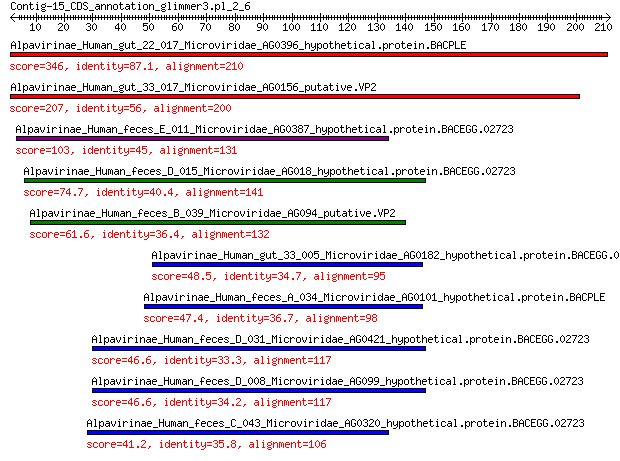
Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.p... 346 2e-120
Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2 207 1e-66
Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.... 103 1e-27
Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.p... 74.7 2e-17
Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2 61.6 3e-13
Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.p... 48.5 1e-08
Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.... 47.4 3e-08
Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.... 46.6 5e-08
Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.p... 46.6 5e-08
Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.... 41.2 2e-06
> Alpavirinae_Human_gut_22_017_Microviridae_AG0396_hypothetical.protein.BACPLE
Length=386
Score = 346 bits (887), Expect = 2e-120, Method: Compositional matrix adjust.
Identities = 183/210 (87%), Positives = 198/210 (94%), Gaps = 0/210 (0%)
Query 1 METLSNIDWGNLTAETRNYLRSTGLARAQLGYVKEQQEVDNMAMTGLIMRAQRSGMLLDN 60
METLSNIDWG LT ETR +L+STGLARAQLGY KEQQE DNMAMTGL++RAQRSGMLLDN
Sbjct 155 METLSNIDWGKLTDETRGFLKSTGLARAQLGYAKEQQEADNMAMTGLVLRAQRSGMLLDN 214
Query 61 EAKGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladealaaartrGQKISN 120
EAKGILNKYLDQ QQLDL+VKAADYYQRM+AGY+SYAE KKALA+EALAAARTRGQ ISN
Sbjct 215 EAKGILNKYLDQHQQLDLSVKAADYYQRMAAGYVSYAEAKKALAEEALAAARTRGQNISN 274
Query 121 EVASRIAESQIAANIAANQSSEAYHNEELKLGLSQDNARSKNIEEWYRSRNEKKRYKYYD 180
EVASRIAESQIAANIAAN+SS AYHNEEL+LGL QDNARSKNIEEWYRSRNEKKRYKY+D
Sbjct 275 EVASRIAESQIAANIAANESSAAYHNEELRLGLPQDNARSKNIEEWYRSRNEKKRYKYFD 334
Query 181 ADKWVEYGTNIGNIIGNFLPRRIVSKTFPR 210
ADKWVEYGT+IGN IGNFLPRR++SK+F R
Sbjct 335 ADKWVEYGTSIGNTIGNFLPRRVISKSFSR 364
> Alpavirinae_Human_gut_33_017_Microviridae_AG0156_putative.VP2
Length=353
Score = 207 bits (526), Expect = 1e-66, Method: Compositional matrix adjust.
Identities = 112/200 (56%), Positives = 152/200 (76%), Gaps = 2/200 (1%)
Query 1 METLSNIDWGNLTAETRNYLRSTGLARAQLGYVKEQQEVDNMAMTGLIMRAQRSGMLLDN 60
M+ LS +DW +T ETR YL++TGLARA+LGY KE QE+DNMA G +++AQ + LL+
Sbjct 155 MQALSQVDWSKMTKETREYLKATGLARARLGYSKEMQELDNMAFAGRLLQAQGTSQLLEA 214
Query 61 EAKGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladealaaartrGQKISN 120
+AK +LN+YLDQ+QQ DLNVKA+ YY +MS G+L+Y + KK +ADE L AR +GQK+SN
Sbjct 215 DAKTVLNRYLDQQQQADLNVKASVYYNQMSQGHLNYNQAKKVIADEILTYARIKGQKLSN 274
Query 121 EVASRIAESQIAANIAANQSSEAYHNEELKLGLSQDNARSKNIEEWYRSRNEKKRYKYYD 180
+VA A+S I A AAN+S+ + E K +++ ARS++IE+WYRSRNE K+YKYYD
Sbjct 275 KVAEATADSLIRATNAANRSNAEFDLEAAK--YNRERARSRSIEDWYRSRNEGKKYKYYD 332
Query 181 ADKWVEYGTNIGNIIGNFLP 200
+DK + YGT+IGN +GNFLP
Sbjct 333 SDKLIHYGTSIGNTVGNFLP 352
> Alpavirinae_Human_feces_E_011_Microviridae_AG0387_hypothetical.protein.BACEGG.02723
Length=410
Score = 103 bits (257), Expect = 1e-27, Method: Compositional matrix adjust.
Identities = 59/131 (45%), Positives = 83/131 (63%), Gaps = 0/131 (0%)
Query 3 TLSNIDWGNLTAETRNYLRSTGLARAQLGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEA 62
TLS IDW T E R +L++TG+ARAQL + +QQ ++NM I RAQR+ +LL N+A
Sbjct 200 TLSGIDWYKFTPEYRAWLQTTGMARAQLSFNTDQQNLENMKWVNKIQRAQRTDILLSNQA 259
Query 63 KGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladealaaartrGQKISNEV 122
KGI+NKYLD Q L L + A +Q ++G LS + K + + + A T G+KISN++
Sbjct 260 KGIINKYLDTSQSLQLKLMANQSFQAFASGRLSLQQCKTEVTKQLMNMAETEGKKISNKI 319
Query 123 ASRIAESQIAA 133
AS A+ I A
Sbjct 320 ASETADQLIGA 330
> Alpavirinae_Human_feces_D_015_Microviridae_AG018_hypothetical.protein.BACEGG.02723
Length=427
Score = 74.7 bits (182), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 57/141 (40%), Positives = 85/141 (60%), Gaps = 0/141 (0%)
Query 6 NIDWGNLTAETRNYLRSTGLARAQLGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKGI 65
+ +W N++ E Y +GL A++G +++ N + ++RA + LLD E+K I
Sbjct 187 DTNWRNVSPEAIRYNIMSGLEAAKIGMENLREQWANQVWSNNLLRANVANSLLDAESKTI 246
Query 66 LNKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladealaaartrGQKISNEVASR 125
LNKYLDQ+QQ DLNVKAA Y + ++ G L E ++ L+ E L AR RG ISN VA++
Sbjct 247 LNKYLDQQQQADLNVKAAHYEELINRGQLHVVEARELLSREVLNYARARGLNISNWVAAK 306
Query 126 IAESQIAANIAANQSSEAYHN 146
A+ + AN AAN +Y+N
Sbjct 307 SAKGLVYANNAANYYEGSYNN 327
> Alpavirinae_Human_feces_B_039_Microviridae_AG094_putative.VP2
Length=380
Score = 61.6 bits (148), Expect = 3e-13, Method: Compositional matrix adjust.
Identities = 48/133 (36%), Positives = 70/133 (53%), Gaps = 1/133 (1%)
Query 8 DWGNLTAETRNYL-RSTGLARAQLGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKGIL 66
+W NL + Y + TG A L E Q + N I AQ + +LL+++A+ I+
Sbjct 158 NWKNLAIGSSGYWNKETGRVSAALDQSTETQNLKNAQFAERISAAQEAQILLNSDAQRIM 217
Query 67 NKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladealaaartrGQKISNEVASRI 126
NKY+DQ QQ DL +KA S G L+ + + + LA+A G+KI N VAS
Sbjct 218 NKYMDQNQQADLFIKAQTLANLQSQGALTEKQIQTEIQRAILASAEASGKKIDNRVASET 277
Query 127 AESQIAANIAANQ 139
A+S I A A+N+
Sbjct 278 ADSLIKAANASNE 290
> Alpavirinae_Human_gut_33_005_Microviridae_AG0182_hypothetical.protein.BACEGG.02723
Length=354
Score = 48.5 bits (114), Expect = 1e-08, Method: Compositional matrix adjust.
Identities = 33/95 (35%), Positives = 57/95 (60%), Gaps = 0/95 (0%)
Query 51 AQRSGMLLDNEAKGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladealaa 110
AQ++ +LL N+A+ +LN YL +E+++ L + A Y+ + G +S + K +A
Sbjct 178 AQKANLLLRNDAQEVLNMYLPEEKRIQLQMNGAQYWNMIREGVISEEQAKNLIASRLEIE 237
Query 111 artrGQKISNEVASRIAESQIAANIAANQSSEAYH 145
ART+GQ ISN++A A+S I A A ++ A++
Sbjct 238 ARTQGQHISNKIAKSTADSIIDATRTAKENEAAFN 272
> Alpavirinae_Human_feces_A_034_Microviridae_AG0101_hypothetical.protein.BACPLE
Length=365
Score = 47.4 bits (111), Expect = 3e-08, Method: Compositional matrix adjust.
Identities = 36/98 (37%), Positives = 54/98 (55%), Gaps = 0/98 (0%)
Query 48 IMRAQRSGMLLDNEAKGILNKYLDQEQQLDLNVKAADYYQRMSAGYLSYaetkkaladea 107
+ AQ++ +LL EA+ ++N YL QE+Q+ L+ A Y+ + G + + K LA
Sbjct 175 VATAQKANLLLSAEAQQVMNMYLPQEKQIQLSTLGAQYWNMIRDGSIKEEQAKNLLATRL 234
Query 108 laaartrGQKISNEVASRIAESQIAANIAANQSSEAYH 145
ART GQ ISN+VA A+S I A A + AY+
Sbjct 235 EIEARTAGQHISNKVARSTADSIIDATNTAKMNEAAYN 272
> Alpavirinae_Human_feces_D_031_Microviridae_AG0421_hypothetical.protein.BACEGG.02723
Length=418
Score = 46.6 bits (109), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 39/119 (33%), Positives = 69/119 (58%), Gaps = 2/119 (2%)
Query 30 LGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKG--ILNKYLDQEQQLDLNVKAADYYQ 87
LG ++ ++N++ I A G LL +A+G I+NK++ +QQ + +K A+ +
Sbjct 203 LGIDLRKKRLENLSTETNIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFA 262
Query 88 RMSAGYLSYaetkkaladealaaartrGQKISNEVASRIAESQIAANIAANQSSEAYHN 146
+ AG LS A+ K + +AL A+ GQK++N +A R+A+ Q A A +++ AY+N
Sbjct 263 QYKAGKLSEAQVKTQIKQQALLEAQAAGQKLNNRLAERLADYQFKAMAAEYRANAAYYN 321
> Alpavirinae_Human_feces_D_008_Microviridae_AG099_hypothetical.protein.BACEGG.02723
Length=418
Score = 46.6 bits (109), Expect = 5e-08, Method: Compositional matrix adjust.
Identities = 40/119 (34%), Positives = 70/119 (59%), Gaps = 2/119 (2%)
Query 30 LGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKG--ILNKYLDQEQQLDLNVKAADYYQ 87
LG ++ ++N++ I A G LL +A+G I+NK++ +QQ + +K A+ +
Sbjct 203 LGIDLRKKRLENLSTETNIKVALAQGALLGLQAEGQRIVNKFMPAQQQAEFFLKTANAFA 262
Query 88 RMSAGYLSYaetkkaladealaaartrGQKISNEVASRIAESQIAANIAANQSSEAYHN 146
+ AG LS A+ K + +AL A+T GQK++N +A R+A+ Q A A +++ AY+N
Sbjct 263 QYKAGKLSEAQVKTQIKQQALLEAQTVGQKLNNRLAERLADYQFKAMAAEYRANAAYYN 321
> Alpavirinae_Human_feces_C_043_Microviridae_AG0320_hypothetical.protein.BACEGG.02723
Length=396
Score = 41.2 bits (95), Expect = 2e-06, Method: Compositional matrix adjust.
Identities = 38/106 (36%), Positives = 54/106 (51%), Gaps = 0/106 (0%)
Query 28 AQLGYVKEQQEVDNMAMTGLIMRAQRSGMLLDNEAKGILNKYLDQEQQLDLNVKAADYYQ 87
AQ+ K E+ N+ A+ + L +A+ LNKYLD QQL L K A+Y
Sbjct 179 AQISIGKGSAEISNLRSQSARNYAETAVASLTADAQRTLNKYLDMGQQLSLITKMAEYSS 238
Query 88 RMSAGYLSYaetkkaladealaaartrGQKISNEVASRIAESQIAA 133
+ L+ A+ + +A+E A GQKISNE+A A+S I A
Sbjct 239 ITAGTELTKAKYRTEIANEIKTLAEANGQKISNEIARSTAQSLIDA 284
Lambda K H a alpha
0.313 0.129 0.359 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 16069636