bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
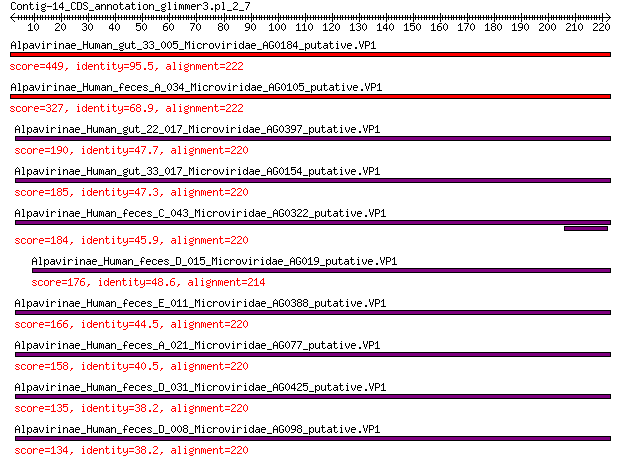
Query= Contig-14_CDS_annotation_glimmer3.pl_2_7
Length=222
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1 449 9e-158
Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1 327 8e-110
Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1 190 5e-58
Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1 185 3e-56
Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1 184 1e-55
Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1 176 7e-53
Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1 166 2e-49
Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1 158 2e-46
Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1 135 2e-38
Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1 134 6e-38
> Alpavirinae_Human_gut_33_005_Microviridae_AG0184_putative.VP1
Length=618
Score = 449 bits (1156), Expect = 9e-158, Method: Compositional matrix adjust.
Identities = 212/222 (95%), Positives = 219/222 (99%), Gaps = 0/222 (0%)
Query 1 MTSHRCQRVCGFDGSIDISAVENTNLSSDEAIIRGKGIGGYRVNKPETFKTTEHGVLMCI 60
+TSHR RVCGFDGSIDISAVENTNLSSDEAIIRGKGIGGYRVNKPETF+TTEHGVLMCI
Sbjct 397 ITSHRSTRVCGFDGSIDISAVENTNLSSDEAIIRGKGIGGYRVNKPETFETTEHGVLMCI 456
Query 61 YHAVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEPRPF 120
YHAVPLLDYAPTGPDLQFMTTVDGDSWPVPE+DS+GFEELPSYSLLNT+ VQPIKEPRPF
Sbjct 457 YHAVPLLDYAPTGPDLQFMTTVDGDSWPVPEMDSIGFEELPSYSLLNTNAVQPIKEPRPF 516
Query 121 GYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGEDYMKIYFDNNNVPGGAHFGFYTWFKVN 180
GYVPRYISWKTSVDVVRGAFIDTLKSWTAPIG+DY+KIYFDNNNVPGGAHFGFYTWFKVN
Sbjct 517 GYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGQDYLKIYFDNNNVPGGAHFGFYTWFKVN 576
Query 181 PSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 222
PSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY
Sbjct 577 PSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 618
> Alpavirinae_Human_feces_A_034_Microviridae_AG0105_putative.VP1
Length=618
Score = 327 bits (837), Expect = 8e-110, Method: Compositional matrix adjust.
Identities = 153/222 (69%), Positives = 173/222 (78%), Gaps = 0/222 (0%)
Query 1 MTSHRCQRVCGFDGSIDISAVENTNLSSDEAIIRGKGIGGYRVNKPETFKTTEHGVLMCI 60
M SHR R+CGFDGSIDISAVENTNL++DEAIIRGKG+GG R+N P F EHGV+MCI
Sbjct 397 MQSHRSTRICGFDGSIDISAVENTNLTADEAIIRGKGLGGQRINDPSDFTCNEHGVIMCI 456
Query 61 YHAVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEPRPF 120
YHA PLLDY PTGPDLQ M+TV G+SWPVPE DS+G E LP SL+N+ + I
Sbjct 457 YHATPLLDYVPTGPDLQLMSTVKGESWPVPEFDSLGMESLPMLSLVNSKAIGDIVARSYA 516
Query 121 GYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGEDYMKIYFDNNNVPGGAHFGFYTWFKVN 180
GYVPRYISWKTS+DVVRGAF DTLKSWTAP+ DYM ++F G+ YTWFKVN
Sbjct 517 GYVPRYISWKTSIDVVRGAFTDTLKSWTAPVDSDYMHVFFGEVIPQEGSPILSYTWFKVN 576
Query 181 PSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 222
PSV+NPIF V DGSWNTDQLL NC FDV+VARNLSYDG+PY
Sbjct 577 PSVLNPIFAVSVDGSWNTDQLLCNCQFDVKVARNLSYDGMPY 618
> Alpavirinae_Human_gut_22_017_Microviridae_AG0397_putative.VP1
Length=607
Score = 190 bits (483), Expect = 5e-58, Method: Compositional matrix adjust.
Identities = 105/227 (46%), Positives = 138/227 (61%), Gaps = 11/227 (5%)
Query 3 SHRCQRVCGFDGSIDISAVENTNLSS--DEAIIRGKGIGGYRVNKPETFKTTEHGVLMCI 60
S+ C + G ++DIS V N NL++ D A+I GKG+G N T+ T EH V+MCI
Sbjct 385 SNMCTYIGGISRNLDISEVVNNNLAAEGDTAVIAGKGVGA--GNGSFTYTTNEHCVVMCI 442
Query 61 YHAVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEPRPF 120
YHAVPLLDY TG D Q + T D +S P+PE D++G E LP + N+ +
Sbjct 443 YHAVPLLDYTITGQDGQLLVT-DAESLPIPEFDNIGMETLPMTQIFNSPKASIVNLFNA- 500
Query 121 GYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGEDYMKIYFDNNNVPGGAHFGF-----YT 175
GY PRY +WKT +DV+ GAF TLKSW +P+ E + +F G + Y
Sbjct 501 GYNPRYFNWKTKLDVINGAFTTTLKSWVSPVTESLLSGWFGFGYSEGDVNSQNKVVLNYK 560
Query 176 WFKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 222
+FKVNPSV++PIFGV AD +W++DQLLVN VARNLS DG+PY
Sbjct 561 FFKVNPSVLDPIFGVAADSTWDSDQLLVNSYIGCYVARNLSRDGVPY 607
> Alpavirinae_Human_gut_33_017_Microviridae_AG0154_putative.VP1
Length=614
Score = 185 bits (470), Expect = 3e-56, Method: Compositional matrix adjust.
Identities = 104/225 (46%), Positives = 138/225 (61%), Gaps = 9/225 (4%)
Query 3 SHRCQRVCGFDGSIDISAVENTNLSSDE--AIIRGKGIGGYRVNKPETFKTTEHGVLMCI 60
S+ C + G ++DIS V N NL+++E A+I GKG+G N T+ T EH V+M I
Sbjct 394 SNLCTYIGGISRNLDISEVVNNNLAAEEDTAVIAGKGVG--TGNGSFTYTTNEHCVIMGI 451
Query 61 YHAVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEPRPF 120
YHAVPLLDY TG D Q + T D +S P+PE D++G E LP + N+S
Sbjct 452 YHAVPLLDYTLTGQDGQLLVT-DAESLPIPEFDNIGLEVLPMAQIFNSSLATAFNLFNA- 509
Query 121 GYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGEDYMK---IYFDNNNVPGGAHFGFYTWF 177
GY PRY +WKT +DV+ GAF TLKSW +P+ E + + +++ G Y +F
Sbjct 510 GYNPRYFNWKTKLDVINGAFTTTLKSWVSPVSESLLSGWARFGASDSKTGTKAVLNYKFF 569
Query 178 KVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 222
KVNPSV++PIFGV AD +W+TDQLLVN V RNLS DG+PY
Sbjct 570 KVNPSVLDPIFGVKADSTWDTDQLLVNSYIGCYVVRNLSRDGVPY 614
> Alpavirinae_Human_feces_C_043_Microviridae_AG0322_putative.VP1
Length=602
Score = 184 bits (466), Expect = 1e-55, Method: Compositional matrix adjust.
Identities = 101/225 (45%), Positives = 136/225 (60%), Gaps = 10/225 (4%)
Query 3 SHRCQRVCGFDGSIDISAVENTNLSSDEAIIRGKGIGGYRVNKPETFKTTEHGVLMCIYH 62
S C+ + G + IS V NTNL S A I+GKG+GG ETF+T EHG+LMCIYH
Sbjct 383 SDHCRYIGGTSSGVTISEVLNTNLESAAADIKGKGVGGSF--GSETFETNEHGILMCIYH 440
Query 63 AVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQP---IKEPRP 119
A P+LDY +G DLQ ++T+ D P+PE D +G E LP +L N + I
Sbjct 441 ATPVLDYLRSGQDLQLLSTLATD-IPIPEFDHIGMEALPIETLFNEQSTEATALINSIPV 499
Query 120 FGYVPRYISWKTSVDVVRGAFIDTLKSWTAP--IGEDYMKIYFDNNNVPGGAHFGFYTWF 177
GY PRYI++KTSVD V GAF TL SW AP + E K+ F+ ++ G + Y +F
Sbjct 500 LGYSPRYIAYKTSVDWVSGAFETTLDSWVAPLTVNEQITKLLFNPDS--GSVYSMNYGFF 557
Query 178 KVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 222
KV P V++PIF +W++DQ LVN F+V+ +NL Y+G+PY
Sbjct 558 KVTPRVLDPIFVQECTDTWDSDQFLVNVSFNVKPVQNLDYNGMPY 602
Score = 21.2 bits (43), Expect = 9.1, Method: Compositional matrix adjust.
Identities = 7/16 (44%), Positives = 13/16 (81%), Gaps = 0/16 (0%)
Query 206 DFDVRVARNLSYDGLP 221
DF++ A+N+S++ LP
Sbjct 182 DFNLLAAKNVSFNVLP 197
> Alpavirinae_Human_feces_D_015_Microviridae_AG019_putative.VP1
Length=584
Score = 176 bits (445), Expect = 7e-53, Method: Compositional matrix adjust.
Identities = 104/228 (46%), Positives = 136/228 (60%), Gaps = 26/228 (11%)
Query 9 VCGFDGSIDISAVENTNLS-SDEAIIRGKGIGGYRVNKPETFKTTEHGVLMCIYHAVPLL 67
+ G S+DIS V NTN++ S+EA+I GKGIG + N E F + GVLMCIYH+VPLL
Sbjct 369 IGGEASSLDISEVVNTNITESNEALIAGKGIGTGQGN--EEFYAKDWGVLMCIYHSVPLL 426
Query 68 DYAPTGPDLQFMTTVDGDSWPVPELDSVGFE------------ELPSYSLLNTSDVQPIK 115
DY + PD Q +++ S+PVPELD++G E ELPS + + +
Sbjct 427 DYVISAPDPQLFASMN-TSFPVPELDAIGLEPITVAYYSNNPIELPSTGGITDAPTTTV- 484
Query 116 EPRPFGYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGEDYMKIYFDNNNVPG-GAHFGFY 174
GY+PRY +WKTS+D V GAF T K W API + G G ++ F
Sbjct 485 -----GYLPRYYAWKTSIDYVLGAFTTTEKEWVAPITPELWSNMLKPLGTKGTGINYNF- 538
Query 175 TWFKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 222
FKVNPS+++PIF V AD W+TD L+N FD+RVARNL YDG+PY
Sbjct 539 --FKVNPSILDPIFAVNADSYWDTDTFLINAAFDIRVARNLDYDGMPY 584
> Alpavirinae_Human_feces_E_011_Microviridae_AG0388_putative.VP1
Length=622
Score = 166 bits (420), Expect = 2e-49, Method: Compositional matrix adjust.
Identities = 98/222 (44%), Positives = 132/222 (59%), Gaps = 6/222 (3%)
Query 3 SHRCQRVCGFDGSIDISAVENTNLSSD--EAIIRGKGIGGYRVNKPETFKTTEHGVLMCI 60
SH Q + G ++DIS V N NL D EA+I GKG+G +++ ++MCI
Sbjct 405 SHMAQYIGGVARNLDISEVVNNNLKDDGSEAVIYGKGVGSGSGKM-RYHTGSQYCIIMCI 463
Query 61 YHAVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIKEPRPF 120
YHA+PLLDYA TG D Q + T D P+PE D++G E +P+ +L N+
Sbjct 464 YHAMPLLDYAITGQDPQLLCTSVED-LPIPEFDNIGMEAVPATTLFNSVLFDGTAVNDFL 522
Query 121 GYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGEDYMKIYFDNNNVPGGAHFGFYTWFKVN 180
GY PRY WK+ +D V GAF TLK W API +DY+ +F N+ G + + +FKVN
Sbjct 523 GYNPRYWPWKSKIDRVHGAFTTTLKDWVAPIDDDYLHNWF--NSKDGKSASISWPFFKVN 580
Query 181 PSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 222
P+ ++ IF VVAD W TDQLL+NCD +V R LS DG+PY
Sbjct 581 PNTLDSIFAVVADSIWETDQLLINCDVSCKVVRPLSQDGMPY 622
> Alpavirinae_Human_feces_A_021_Microviridae_AG077_putative.VP1
Length=622
Score = 158 bits (399), Expect = 2e-46, Method: Compositional matrix adjust.
Identities = 89/238 (37%), Positives = 128/238 (54%), Gaps = 23/238 (10%)
Query 3 SHRCQRVCGFDGSIDISAVENTNLSSDE--------AIIRGKGI--GGYRVNKPETFKTT 52
S+ Q + G +DIS V N NL+ A+I GKG+ G VN +
Sbjct 390 SYMSQYIGGQFAQMDISEVVNQNLTDQAGSDAAQYPALIAGKGVNSGDGNVN----YTAR 445
Query 53 EHGVLMCIYHAVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQ 112
+HG++M IYHAVPLLDY TG D + T + W +PE D++G + LP +L N++ V
Sbjct 446 QHGIIMGIYHAVPLLDYERTGQDQDLLIT-SAEEWAIPEFDAIGMQTLPLGTLFNSNKVS 504
Query 113 PIKEPR------PFGYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGEDYMKIYFDN--NN 164
+ R P GYVPRY++WKT +D + GAF + K+W API D++ + N +N
Sbjct 505 GDSDFRLHGAAYPIGYVPRYVNWKTDIDEIFGAFRSSEKTWVAPIDADFITNWVKNVADN 564
Query 165 VPGGAHFGFYTWFKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 222
Y WFKVNP++++ IF V AD + +TD ++ RNL Y G+PY
Sbjct 565 ASAVQSLFNYNWFKVNPAILDDIFAVKADSTMDTDTFKTRMMMSIKCVRNLDYSGMPY 622
> Alpavirinae_Human_feces_D_031_Microviridae_AG0425_putative.VP1
Length=614
Score = 135 bits (341), Expect = 2e-38, Method: Compositional matrix adjust.
Identities = 84/230 (37%), Positives = 121/230 (53%), Gaps = 16/230 (7%)
Query 3 SHRCQRVCGFDGSIDISAVENTNLSSDE-AIIRGKGIGGYRVNKPETFKTTEHGVLMCIY 61
S +C + G +ID+S V NTN++ D A I GKG+G + + F E+G+++ IY
Sbjct 391 SDKCVYLGGSSSNIDLSEVVNTNITGDNVAEIAGKGVGTGQGSFSGQFD--EYGIIIGIY 448
Query 62 HAVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIK----EP 117
H VPLLDY TG + T D P PE DS+G + + +N+ V
Sbjct 449 HNVPLLDYVITGQPQNLLYTNTAD-LPFPEFDSIGMQTIQFGRFVNSKSVSWTSGVDYRV 507
Query 118 RPFGYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGEDYMKIYFDNNNVPGGA-----HFG 172
+ GY+PR+ KT D V GAF TLK+W AP+ Y+ + ++ G ++G
Sbjct 508 QTMGYLPRFFDVKTRYDEVLGAFRSTLKNWVAPLDPSYVSKWLQSSVTSSGKLALNLNYG 567
Query 173 FYTWFKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 222
F FKVNP V++ IF V D + +TDQ L D++ RN YDG+PY
Sbjct 568 F---FKVNPRVLDSIFNVKCDSTIDTDQFLTALYMDIKAVRNFDYDGMPY 614
> Alpavirinae_Human_feces_D_008_Microviridae_AG098_putative.VP1
Length=618
Score = 134 bits (337), Expect = 6e-38, Method: Compositional matrix adjust.
Identities = 84/227 (37%), Positives = 118/227 (52%), Gaps = 10/227 (4%)
Query 3 SHRCQRVCGFDGSIDISAVENTNLSSDE-AIIRGKGIGGYRVNKPETFKTTEHGVLMCIY 61
S +C + G +ID+S V NTN++ D A I GKG+G + + F T +G++M IY
Sbjct 395 SDKCMYLGGSSSNIDLSEVVNTNITGDNIAEIAGKGVGTGQGSFSGNFDT--YGIIMGIY 452
Query 62 HAVPLLDYAPTGPDLQFMTTVDGDSWPVPELDSVGFEELPSYSLLNTSDVQPIK----EP 117
H VPLLDY TG + T D P PE DS+G + + +N+ V
Sbjct 453 HNVPLLDYVITGQPQNLLYTNTAD-LPFPEYDSIGMQTIQFGRFVNSKAVGWTSGVDYRT 511
Query 118 RPFGYVPRYISWKTSVDVVRGAFIDTLKSWTAPIGEDYMKIYFDNNNVPGGAHFGF--YT 175
+ GY+PR+ KT D V GAF TLK+W AP+ + + + G F Y
Sbjct 512 QTMGYLPRFFDVKTRYDEVLGAFRSTLKNWVAPLDPANLPQWLQTSVTSSGKLFLNLNYG 571
Query 176 WFKVNPSVVNPIFGVVADGSWNTDQLLVNCDFDVRVARNLSYDGLPY 222
+FKVNP V++ IF V D + +TDQ L D++ RN YDG+PY
Sbjct 572 FFKVNPRVLDSIFNVKCDSTIDTDQFLTTLYMDIKAVRNFDYDGMPY 618
Lambda K H a alpha
0.320 0.140 0.447 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 17280362