bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: Microviridae_proteins.fasta
575 sequences; 147,441 total letters
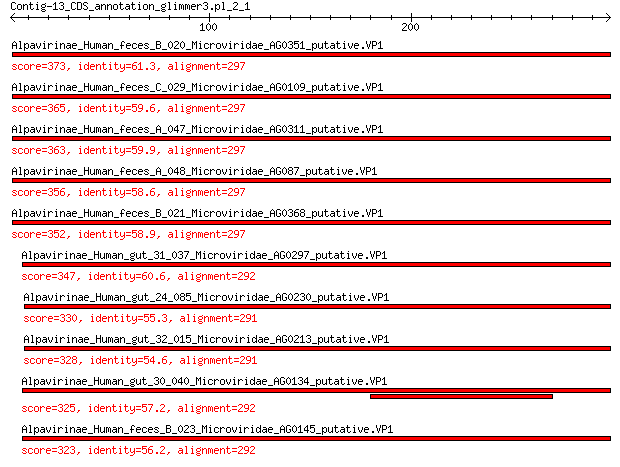
Query= Contig-13_CDS_annotation_glimmer3.pl_2_1
Length=298
Score E
Sequences producing significant alignments: (Bits) Value
Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1 373 4e-126
Alpavirinae_Human_feces_C_029_Microviridae_AG0109_putative.VP1 365 2e-123
Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1 363 2e-122
Alpavirinae_Human_feces_A_048_Microviridae_AG087_putative.VP1 356 1e-119
Alpavirinae_Human_feces_B_021_Microviridae_AG0368_putative.VP1 352 3e-118
Alpavirinae_Human_gut_31_037_Microviridae_AG0297_putative.VP1 347 5e-116
Alpavirinae_Human_gut_24_085_Microviridae_AG0230_putative.VP1 330 8e-110
Alpavirinae_Human_gut_32_015_Microviridae_AG0213_putative.VP1 328 7e-109
Alpavirinae_Human_gut_30_040_Microviridae_AG0134_putative.VP1 325 3e-107
Alpavirinae_Human_feces_B_023_Microviridae_AG0145_putative.VP1 323 6e-107
> Alpavirinae_Human_feces_B_020_Microviridae_AG0351_putative.VP1
Length=651
Score = 373 bits (957), Expect = 4e-126, Method: Compositional matrix adjust.
Identities = 182/308 (59%), Positives = 229/308 (74%), Gaps = 11/308 (4%)
Query 2 LRTKQPMYGLAVKTYQSDIFNNWIQTEWLEGENGINAITAVDTSIGSFTMDTLNLAKKVY 61
L+T +P YGL +KTY SD++ NWI T+W++G NGIN I++VD S G TMD LNLA+KVY
Sbjct 344 LKTTRPQYGLLLKTYNSDLYQNWINTDWIDGANGINEISSVDVSEGKLTMDALNLAQKVY 403
Query 62 NMLNRIAVSDGTYKSWMETVF-SAKYVERTETPIYYGGMSQEIIFEEVVSTSATGEEPLG 120
NMLNRIAVS GTY+ W+ETV+ S +Y+ER ETP + GG SQEI+F+EVVS SAT +EPLG
Sbjct 404 NMLNRIAVSGGTYRDWLETVYASGQYIERCETPTFEGGTSQEIVFQEVVSNSATEDEPLG 463
Query 121 TLAGKGRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNWETMNDLHKPAL 180
TLAG+G A +KGGK+ ++ EP YI+ I SITPR+DYSQGN+F +W+T++D+HKPAL
Sbjct 464 TLAGRGVNAGKQKGGKIKVRATEPGYIMCITSITPRIDYSQGNDFDTDWKTLDDMHKPAL 523
Query 181 DMIGYQDLTMEKAAWWTEEHT--SDTEFAQKSIGKTVAWVDYMTNYNKNYGNFASGENEN 238
D IGYQD AWW + +T +T + + GKTVAW+DYMTN NK YGNFA+G +E
Sbjct 524 DGIGYQDSVNTGRAWWDDVYTGAQETNLVKHTAGKTVAWIDYMTNVNKTYGNFAAGMSEA 583
Query 239 FMTLDRNYNVE-----NP---DFTTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRKI 290
FM L+RNY ++ NP D TTYIDP KYN IFAD S +MNFWVQI + RR +
Sbjct 584 FMVLNRNYEIKYDSGTNPRIADLTTYIDPIKYNYIFADTSIDAMNFWVQIKFDITARRLM 643
Query 291 SAKSIPNL 298
SAK IPNL
Sbjct 644 SAKQIPNL 651
> Alpavirinae_Human_feces_C_029_Microviridae_AG0109_putative.VP1
Length=640
Score = 365 bits (938), Expect = 2e-123, Method: Compositional matrix adjust.
Identities = 177/309 (57%), Positives = 227/309 (73%), Gaps = 12/309 (4%)
Query 2 LRTKQPMYGLAVKTYQSDIFNNWIQTEWLEGENGINAITAVDTSIGSFTMDTLNLAKKVY 61
L+T P +GLA+KTY SD++ NWI T+W+EG NGIN I++VD S G+ +MD LNLA+KVY
Sbjct 332 LKTIGPQFGLALKTYNSDLYQNWINTDWIEGVNGINEISSVDVSDGNLSMDALNLAQKVY 391
Query 62 NMLNRIAVSDGTYKSWMETVFSA-KYVERTETPIYYGGMSQEIIFEEVVSTSATGEEPLG 120
NMLNRIAVS GTY+ W+ETVF+ +Y+ER ETP++ GG SQEI+F+EV+S SAT +EPLG
Sbjct 392 NMLNRIAVSGGTYRDWLETVFTGGEYMERCETPVFEGGTSQEIVFQEVISNSATEQEPLG 451
Query 121 TLAGKGRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNWETMNDLHKPAL 180
TLAG+G ++GG + IK+ EP YI+ ICSITPR+DYSQGN + +TM+DLHKPAL
Sbjct 452 TLAGRGITTNKQRGGHIKIKVTEPGYIMCICSITPRIDYSQGNQWDTELKTMDDLHKPAL 511
Query 181 DMIGYQDLTMEKAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKNYGNFASGENENFM 240
D IGYQD + AWW + +D + + + GKTVAW++YMTN N+ YGNFA +NE FM
Sbjct 512 DGIGYQDSINSERAWWAGYYNADPDKKETAAGKTVAWINYMTNVNRTYGNFAIKDNEAFM 571
Query 241 TLDRNYNVE-----NP------DFTTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRK 289
++RNY +E P D +TYIDPAK+N IFAD S +MNFWVQ + K RR
Sbjct 572 VMNRNYELEITGGSTPTKITIGDLSTYIDPAKFNYIFADTSLEAMNFWVQTKFDIKARRL 631
Query 290 ISAKSIPNL 298
ISAK IPNL
Sbjct 632 ISAKQIPNL 640
> Alpavirinae_Human_feces_A_047_Microviridae_AG0311_putative.VP1
Length=637
Score = 363 bits (931), Expect = 2e-122, Method: Compositional matrix adjust.
Identities = 178/309 (58%), Positives = 224/309 (72%), Gaps = 12/309 (4%)
Query 2 LRTKQPMYGLAVKTYQSDIFNNWIQTEWLEGENGINAITAVDTSIGSFTMDTLNLAKKVY 61
L+T P +GLA+KTY SD++ NWI T+W+EG NGIN I++VD S GS +MD LNLA+KVY
Sbjct 329 LKTTAPQFGLALKTYNSDLYQNWINTDWIEGVNGINEISSVDVSDGSLSMDALNLAQKVY 388
Query 62 NMLNRIAVSDGTYKSWMETVFSA-KYVERTETPIYYGGMSQEIIFEEVVSTSATGEEPLG 120
NMLNRIAVS GTY+ W+ETVF+ +Y+ER ETPI+ GG SQEIIF+EV+S SAT +EPLG
Sbjct 389 NMLNRIAVSGGTYRDWLETVFTGGEYMERCETPIFEGGTSQEIIFQEVISNSATEQEPLG 448
Query 121 TLAGKGRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNWETMNDLHKPAL 180
TLAG+G ++GG V IK+ EP YI+ ICSITPR+DYSQGN + +TM+DLHKPAL
Sbjct 449 TLAGRGITTNKQRGGHVKIKVTEPGYIMCICSITPRIDYSQGNQWDTELKTMDDLHKPAL 508
Query 181 DMIGYQDLTMEKAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKNYGNFASGENENFM 240
D IGYQD + AWW ++ + + GKTVAW++YMTN N+ YGNFA +NE FM
Sbjct 509 DGIGYQDSINSERAWWAGYYSQGPLKVETAAGKTVAWINYMTNVNRTYGNFAIKDNEAFM 568
Query 241 TLDRNYNVE---NP--------DFTTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRK 289
++RNY ++ P D +TYIDP K+N IFAD S +MNFWVQ + K RR
Sbjct 569 VMNRNYELQINGGPTPTSIRIGDLSTYIDPVKFNYIFADTSLEAMNFWVQTKFDIKARRL 628
Query 290 ISAKSIPNL 298
ISAK IPNL
Sbjct 629 ISAKQIPNL 637
> Alpavirinae_Human_feces_A_048_Microviridae_AG087_putative.VP1
Length=650
Score = 356 bits (913), Expect = 1e-119, Method: Compositional matrix adjust.
Identities = 174/308 (56%), Positives = 219/308 (71%), Gaps = 12/308 (4%)
Query 2 LRTKQPMYGLAVKTYQSDIFNNWIQTEWLEGENGINAITAVDTSIGSFTMDTLNLAKKVY 61
L T YGL +KTY SD+ NWI TEW++G GIN I+AVD + G TMD LNL++K+Y
Sbjct 344 LNTSSVQYGLCLKTYNSDLLQNWINTEWIDGVAGINEISAVDVTDGKLTMDALNLSQKIY 403
Query 62 NMLNRIAVSDGTYKSWMETVFSA-KYVERTETPIYYGGMSQEIIFEEVVSTSATGEEPLG 120
NMLNRIAVS GTY+ W+ETVF+ Y+ER ETP++ GGMS E++F+EV+S SA+GE+PLG
Sbjct 404 NMLNRIAVSGGTYRDWLETVFTGGNYMERCETPMFEGGMSTEVVFQEVISNSASGEQPLG 463
Query 121 TLAGKGRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNWETMNDLHKPAL 180
TLAG+G +KGG + IK+ EP +I+GI SITPR+DYSQGN F +T++D+HKPAL
Sbjct 464 TLAGRGYDTGKQKGGHIKIKVTEPCFIMGIGSITPRIDYSQGNEFFNELQTVDDIHKPAL 523
Query 181 DMIGYQDLTMEKAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKNYGNFASGENENFM 240
D IGYQD + AWW + D Q S GKTVAW++YMTN N+ +GNFA +NE FM
Sbjct 524 DGIGYQDSLNWQRAWWDDTRMQDNGRIQSSAGKTVAWINYMTNINRTFGNFAINDNEAFM 583
Query 241 TLDRNYNVENP----------DFTTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRKI 290
L+RNY + NP D TTYIDP K+N IFAD + +MNFWVQ + KVRR I
Sbjct 584 VLNRNYEL-NPNAGTNETKIADLTTYIDPVKFNYIFADTNLDAMNFWVQTKFDIKVRRLI 642
Query 291 SAKSIPNL 298
SAK IPNL
Sbjct 643 SAKQIPNL 650
> Alpavirinae_Human_feces_B_021_Microviridae_AG0368_putative.VP1
Length=649
Score = 352 bits (904), Expect = 3e-118, Method: Compositional matrix adjust.
Identities = 175/308 (57%), Positives = 219/308 (71%), Gaps = 12/308 (4%)
Query 2 LRTKQPMYGLAVKTYQSDIFNNWIQTEWLEGENGINAITAVDTSIGSFTMDTLNLAKKVY 61
L T YGL +KTY SD+ NWI TEW++G GIN I+AVD + G TMD LNLA+KVY
Sbjct 343 LNTASVQYGLCLKTYNSDLLQNWINTEWIDGVTGINEISAVDVTDGKLTMDALNLAQKVY 402
Query 62 NMLNRIAVSDGTYKSWMETVFSA-KYVERTETPIYYGGMSQEIIFEEVVSTSATGEEPLG 120
NMLNRIAVS GTY+ W+ETVF+ Y+ER ETP++ GGMS EI+F+EV+S SA+GE+PLG
Sbjct 403 NMLNRIAVSGGTYRDWLETVFTGGNYMERCETPMFEGGMSTEIVFQEVISNSASGEQPLG 462
Query 121 TLAGKGRLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNWETMNDLHKPAL 180
TLAG+G +KGG + IK+ EP +I+GI SITPR+DYSQGN F +T++D+HKPAL
Sbjct 463 TLAGRGYDTGKQKGGHIKIKVTEPCFIMGIGSITPRIDYSQGNEFYNELKTVDDIHKPAL 522
Query 181 DMIGYQDLTMEKAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKNYGNFASGENENFM 240
D IGYQD + AWW + + Q S GKTVAW++YMTN N+ +GNFA +NE FM
Sbjct 523 DGIGYQDSLNWQRAWWDDTRMENNGRIQSSAGKTVAWINYMTNINRTFGNFAINDNEAFM 582
Query 241 TLDRNYNVENP----------DFTTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRKI 290
L+RNY + NP D TTYIDP K+N IFA ++ +MNFWVQ + KVRR I
Sbjct 583 VLNRNYEL-NPNAGTNETKIADLTTYIDPVKFNYIFAYKNLDAMNFWVQTKFDIKVRRLI 641
Query 291 SAKSIPNL 298
SAK IPNL
Sbjct 642 SAKQIPNL 649
> Alpavirinae_Human_gut_31_037_Microviridae_AG0297_putative.VP1
Length=668
Score = 347 bits (891), Expect = 5e-116, Method: Compositional matrix adjust.
Identities = 177/298 (59%), Positives = 219/298 (73%), Gaps = 9/298 (3%)
Query 7 PMYGLAVKTYQSDIFNNWIQTEWLEGENGINAITAVDTSIGSFTMDTLNLAKKVYNMLNR 66
P +GL KTYQSD+ NWI TEWL+G++GIN ITAV G FT+D LNLA KVYN LNR
Sbjct 374 PEWGLVTKTYQSDLCQNWINTEWLDGDDGINQITAVAVEDGKFTIDALNLANKVYNYLNR 433
Query 67 IAVSDGTYKSWMETVFSAKYVERTETPIYYGGMSQEIIFEEVVSTSATGEEPLGTLAGKG 126
IA+SDG+Y+SW+ET ++ Y ERTETPIY GG S EIIF+EVVSTSA +EPLG+LAG+G
Sbjct 434 IAISDGSYRSWLETTWTGSYTERTETPIYCGGSSAEIIFQEVVSTSAATDEPLGSLAGRG 493
Query 127 RLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNWETMNDLHKPALDMIGYQ 186
+ N KGG V ++ EP Y+IGI SITPRLDY+QGN + +N ++++DLHKPALD IG+Q
Sbjct 494 TDS-NHKGGYVTVRATEPGYLIGITSITPRLDYTQGNQWDVNLDSIDDLHKPALDGIGFQ 552
Query 187 DLTME-KAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKNYGNFASGENENFMTLDRN 245
DL+ E A T+ +T + QK IGK AW+DYMTN N+ YGNF + NENFM L R
Sbjct 553 DLSAELLHAGTTQINTINDTITQKFIGKQPAWIDYMTNINRAYGNFRT--NENFMILSRQ 610
Query 246 YNVEN-----PDFTTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRKISAKSIPNL 298
Y+++ D TTYIDP YNGIFADQS + NFWVQIG+ + RR +SAK IPNL
Sbjct 611 YSLDYKKNTIKDMTTYIDPDLYNGIFADQSFDAQNFWVQIGIEIEARRLMSAKIIPNL 668
> Alpavirinae_Human_gut_24_085_Microviridae_AG0230_putative.VP1
Length=653
Score = 330 bits (847), Expect = 8e-110, Method: Compositional matrix adjust.
Identities = 161/299 (54%), Positives = 214/299 (72%), Gaps = 8/299 (3%)
Query 8 MYGLAVKTYQSDIFNNWIQTEWLEGENGINAITAVDTSIGSFTMDTLNLAKKVYNMLNRI 67
M GLAVKTY SD+F NW+ TE +EG GIN ++V +MD LNLA+KVY+ LNRI
Sbjct 355 MAGLAVKTYDSDVFQNWVNTELIEGAKGINEASSVAIIDNKLSMDALNLAQKVYDFLNRI 414
Query 68 AVSDGTYKSWMETVFSA-KYVERTETPIYYGGMSQEIIFEEVVSTSATGEEPLGTLAGKG 126
AVS TYK W+ET ++A Y+ER ETP++ GGM+Q I F+EVVS + T +EPLG+LAG+G
Sbjct 415 AVSGNTYKDWLETAYTAGNYIERPETPLFEGGMTQLIEFQEVVSNAGTEQEPLGSLAGRG 474
Query 127 RLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTM-NWETMNDLHKPALDMIGY 185
K G++ +KI EPSYI+GI +ITP +DYSQGN++ M N + M+D HKPA D IG+
Sbjct 475 VTTQQKGDGEIYMKISEPSYIMGIVAITPMIDYSQGNDWDMTNIKNMDDFHKPAFDGIGF 534
Query 186 QDLTMEKAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKNYGNFASGENENFMTLDRN 245
+D E+ A+WT E+ + + + GKTVAW++YMTN+NK +G FA+GE+E+FM ++RN
Sbjct 535 EDSMNEQRAYWTAEYNNGEKISDTKAGKTVAWINYMTNFNKTFGEFAAGESEDFMVMNRN 594
Query 246 YNVEN------PDFTTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRKISAKSIPNL 298
Y + D +TYIDP+KYN IFAD+S S+ NFWVQ V +VRR ISAK IPNL
Sbjct 595 YERDEENDSLISDLSTYIDPSKYNQIFADESLSAQNFWVQTAVQMEVRRNISAKQIPNL 653
> Alpavirinae_Human_gut_32_015_Microviridae_AG0213_putative.VP1
Length=653
Score = 328 bits (841), Expect = 7e-109, Method: Compositional matrix adjust.
Identities = 159/299 (53%), Positives = 212/299 (71%), Gaps = 8/299 (3%)
Query 8 MYGLAVKTYQSDIFNNWIQTEWLEGENGINAITAVDTSIGSFTMDTLNLAKKVYNMLNRI 67
M GLAVKTY SD+F NW+ TE ++G GIN ++V +MD LNLA+KVY+ LNRI
Sbjct 355 MAGLAVKTYDSDVFQNWVNTELIDGAKGINEASSVAIVDNKLSMDALNLAQKVYDFLNRI 414
Query 68 AVSDGTYKSWMETVFSA-KYVERTETPIYYGGMSQEIIFEEVVSTSATGEEPLGTLAGKG 126
AVS TYK W+ET ++A Y+ER ETP++ GGM+Q I F+EV+S + T +EPLGTLAG+G
Sbjct 415 AVSGNTYKDWLETAYTAGNYIERPETPLFEGGMTQLIEFQEVISNAGTEQEPLGTLAGRG 474
Query 127 RLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTM-NWETMNDLHKPALDMIGY 185
+ G++ +KI EPSYIIGI +ITP +DYSQGN++ M N + M+D HKPA D IG+
Sbjct 475 VTTQQRGDGEIYMKISEPSYIIGIVAITPMIDYSQGNDWDMTNIKNMDDFHKPAFDGIGF 534
Query 186 QDLTMEKAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKNYGNFASGENENFMTLDRN 245
+D E+ A+WT E++ + + GKTVAW++YMTN+NK +G FA+GE+E+FM ++RN
Sbjct 535 EDSMNEQRAYWTAEYSDGEKISDTKAGKTVAWINYMTNFNKTFGEFAAGESEDFMVMNRN 594
Query 246 YNVEN------PDFTTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRKISAKSIPNL 298
Y + D +TYIDP+KYN IFAD S S+ NFWVQ +VRR ISAK IPNL
Sbjct 595 YERDEDDDSLISDLSTYIDPSKYNQIFADVSLSAQNFWVQTAAQIEVRRNISAKQIPNL 653
> Alpavirinae_Human_gut_30_040_Microviridae_AG0134_putative.VP1
Length=679
Score = 325 bits (832), Expect = 3e-107, Method: Compositional matrix adjust.
Identities = 167/293 (57%), Positives = 209/293 (71%), Gaps = 9/293 (3%)
Query 7 PMYGLAVKTYQSDIFNNWIQTEWLEGENGINAITAVDTSIGSFTMDTLNLAKKVYNMLNR 66
PM GLA+KTYQSDI NW+ TEW++GE GIN+ITA+DTS GSFT+DTLNLAKKVY MLNR
Sbjct 395 PMVGLALKTYQSDINTNWVNTEWIDGETGINSITAIDTSSGSFTLDTLNLAKKVYTMLNR 454
Query 67 IAVSDGTYKSWMETVFSAKYVERTETPIYYGGMSQEIIFEEVVSTSATGEEPLGTLAGKG 126
IAVSDG+Y +W++TV+++ + ETP+Y GG S EI F+EV++ S T ++PLG LAG+G
Sbjct 455 IAVSDGSYNAWIQTVYTSGGLNHIETPLYLGGSSLEIEFQEVINNSGTEDQPLGALAGRG 514
Query 127 RLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTMNWETMNDLHKPALDMIGYQ 186
+A N KGG +V K DEP YI I SITPR+DY QGN + + E+++DLHKP LD IG+Q
Sbjct 515 -VATNHKGGNIVFKADEPGYIFCITSITPRVDYFQGNEWDLYLESLDDLHKPQLDGIGFQ 573
Query 187 DLTMEKAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKNYGNFASGENENFMTLDRNY 246
D + + E SIGK AWV YMTN NK YGNFA ENE +M L+R +
Sbjct 574 D-----RLYRHLNSSCVREDLNISIGKQPAWVQYMTNVNKTYGNFALVENEGWMCLNRIF 628
Query 247 NVENPD-FTTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRKISAKSIPNL 298
EN D +TTYI P YN IFAD ++ NFWVQI N+K RR +SAK IPN+
Sbjct 629 --ENTDTYTTYIQPHLYNNIFADTDLTAQNFWVQIAFNYKPRRVMSAKIIPNI 679
Score = 22.7 bits (47), Expect = 4.3, Method: Compositional matrix adjust.
Identities = 25/98 (26%), Positives = 41/98 (42%), Gaps = 25/98 (26%)
Query 180 LDMIGYQDLTMEKAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKNYGNFASGENENF 239
L +GY+ E WT +I K + + D NY +A+ + E+F
Sbjct 145 LSYLGYKSFNKETGVDWTW-----------NITKALMYYDIFKNY------YANKQEESF 187
Query 240 MTLD-----RNYNV-ENPD--FTTYIDPAKYNGIFADQ 269
T+D ++ N+ NP+ F ID NG++ Q
Sbjct 188 YTIDGARELKSSNLSSNPNTYFIAAIDGPDVNGLYRIQ 225
> Alpavirinae_Human_feces_B_023_Microviridae_AG0145_putative.VP1
Length=668
Score = 323 bits (829), Expect = 6e-107, Method: Compositional matrix adjust.
Identities = 164/306 (54%), Positives = 218/306 (71%), Gaps = 23/306 (8%)
Query 7 PMYGLAVKTYQSDIFNNWIQTEWLEGENGINAITAVDTSIGSFTMDTLNLAKKVYNMLNR 66
P GL +KTYQSDIF NWI +EW++GENGI+AITA+ T FT+D LNL+KKVY+MLNR
Sbjct 372 PQCGLLLKTYQSDIFTNWINSEWIDGENGISAITAISTEGNKFTIDQLNLSKKVYDMLNR 431
Query 67 IAVSDGTYKSWMETVFSAKYVERTETPIYYGGMSQEIIFEEVVSTSATGEEPLGTLAGKG 126
IA+S GTY+ W+ETV+++++ TETP+Y GGMS EI F+EVVS SAT EEPLGTLAG+G
Sbjct 432 IAISGGTYQDWVETVYTSEWNMHTETPVYEGGMSAEIEFQEVVSNSATEEEPLGTLAGRG 491
Query 127 RLAPNKKGGKVVIKIDEPSYIIGICSITPRLDYSQGNNFTM-NWETMNDLHKPALDMIGY 185
A NKKGG++ IK+ EP YI+GI SITPR+DY QGN++ + + +TM+D+HKP LD IGY
Sbjct 492 -FASNKKGGQLHIKVTEPCYIMGIASITPRVDYCQGNDWDITSLDTMDDIHKPQLDSIGY 550
Query 186 QDLTMEKAAWWTEEHTSDTEFAQKSIGKTVAWVDYMTNYNKNYGNFAS--GENENFMTLD 243
QDL E+ + ++GK +W++YMT++NK YG FA+ GE E FM L+
Sbjct 551 QDLMQEQMNAQASRNL--------AVGKQPSWINYMTSFNKTYGTFANEDGEGEAFMVLN 602
Query 244 RNYNVENPDF-----------TTYIDPAKYNGIFADQSRSSMNFWVQIGVNWKVRRKISA 292
R ++++ D +TYIDP++YN IFA+ SMNFWVQ+G + RR +SA
Sbjct 603 RYFDIKEIDAGTETGVKVYNTSTYIDPSQYNYIFAETGTKSMNFWVQLGFGIEARRVMSA 662
Query 293 KSIPNL 298
IPNL
Sbjct 663 SQIPNL 668
Lambda K H a alpha
0.315 0.131 0.395 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 24918430