bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-8_CDS_annotation_glimmer3.pl_2_4
Length=156
Score E
Sequences producing significant alignments: (Bits) Value
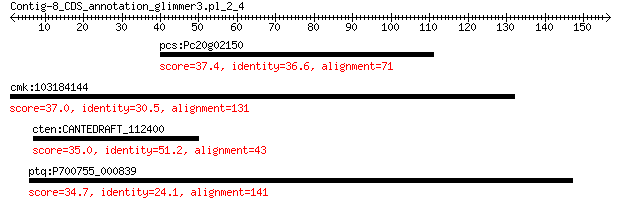
pcs:Pc20g02150 Pc20g02150 37.4 0.86
cmk:103184144 tenm2; teneurin transmembrane protein 2 37.0
cten:CANTEDRAFT_112400 MFS general substrate transporter 35.0 5.9
ptq:P700755_000839 lysyl-tRNA synthetase (class II) LysS 34.7 7.9
> pcs:Pc20g02150 Pc20g02150
Length=551
Score = 37.4 bits (85), Expect = 0.86, Method: Compositional matrix adjust.
Identities = 26/77 (34%), Positives = 42/77 (55%), Gaps = 8/77 (10%)
Query 40 ADSDGVLHVLNDISLL------FNQQRLENRLSSTELREMFNRYSPNKSRYLAQLDDDTL 93
A D LH +++SL F+++ L+ L EL E+ R+ ++R+LA D
Sbjct 227 AHEDVPLHASDEVSLPRLEIPRFSKEELQRSLDDPELSELHERHQMERNRHLA--FHDAA 284
Query 94 LSTLKSRHIQSLSEIKS 110
LSTL+ RH ++SE +S
Sbjct 285 LSTLRRRHQTAVSEHQS 301
> cmk:103184144 tenm2; teneurin transmembrane protein 2
Length=2757
Score = 37.0 bits (84), Expect = 1.5, Method: Compositional matrix adjust.
Identities = 40/137 (29%), Positives = 65/137 (47%), Gaps = 14/137 (10%)
Query 1 MSRKVKKERYSSRLSYNYDY--DSKKSKFVVRDKLAVLSSYADSDGVLHVLND---ISLL 55
+ R++K Y++ YNYDY D + V DK SY D +G LH+LN I L+
Sbjct 2169 IKREIKVGPYANTTKYNYDYDGDGQLQTVTVNDKATWRYSY-DLNGNLHLLNPGSSIRLM 2227
Query 56 FNQQRLENRLSSTELREMFNRYSPNKSRYLAQLDDDTLLSTLKSRHIQSLSEIKSW-TEY 114
+ L +R+ T L ++ +Y ++ L+Q D KS I+ S++ W +Y
Sbjct 2228 PLRYDLRDRI--TRLGDV--QYKLDEDGLLSQRGSDVFEYNSKSLLIRVYSKVNGWNVQY 2283
Query 115 CIENFDSLLKAQQAKEN 131
+D L + +K N
Sbjct 2284 ---RYDGLARRVSSKTN 2297
> cten:CANTEDRAFT_112400 MFS general substrate transporter
Length=470
Score = 35.0 bits (79), Expect = 5.9, Method: Composition-based stats.
Identities = 22/44 (50%), Positives = 28/44 (64%), Gaps = 2/44 (5%)
Query 7 KERYSSRLSYNYDYDSKKSKFV-VRDKLAVLSSYADSDGVLHVL 49
KER++ +S N DYDSK+ KFV V KL L+S DS + VL
Sbjct 243 KERFTKSVS-NDDYDSKRHKFVGVSKKLFNLNSLTDSKFIFLVL 285
> ptq:P700755_000839 lysyl-tRNA synthetase (class II) LysS
Length=563
Score = 34.7 bits (78), Expect = 7.9, Method: Composition-based stats.
Identities = 34/141 (24%), Positives = 64/141 (45%), Gaps = 11/141 (8%)
Query 6 KKERYSSRLSYNYDYDSKKSKFVVRDKLAVLSSYADSDGVLHVLNDISLLFNQQRLENRL 65
++ER+ ++ + D + ++F+ D L L L + D ++F L N
Sbjct 428 QRERFEDQMKLSQKGDDEATEFIDYDFLRALEYGMPPTSGLGIGMDRLIMF----LTNNP 483
Query 66 SSTELREMFNRYSPNKSRYLAQLDDDTLLSTLKSRHIQSLSEIKSWTEYCIENFDSLLKA 125
S E+ F + P K + ++ ++ LK Q+L EIKS +E + +D +K
Sbjct 484 SIQEVL-FFPQMRPEKKKVELTEEEKVVIDLLKVTSPQALEEIKSKSELSNKKWDKAIKG 542
Query 126 QQAKENEIDDIQETVDEPSAG 146
++K+ Q TVD+ + G
Sbjct 543 LRSKD------QATVDKTNEG 557
Lambda K H a alpha
0.310 0.126 0.336 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 126812206336