bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-8_CDS_annotation_glimmer3.pl_2_1
Length=461
Score E
Sequences producing significant alignments: (Bits) Value
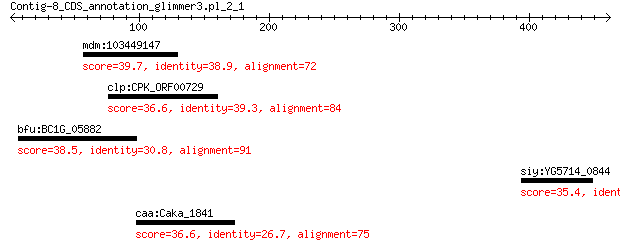
mdm:103449147 ARF guanine-nucleotide exchange factor GNL2-like 39.7 1.7
clp:CPK_ORF00729 hypothetical protein 36.6 2.4
bfu:BC1G_05882 hypothetical protein 38.5 3.0
siy:YG5714_0844 hypothetical protein 35.4 5.0
caa:Caka_1841 TetR family transcriptional regulator 36.6 6.7
> mdm:103449147 ARF guanine-nucleotide exchange factor GNL2-like
Length=1384
Score = 39.7 bits (91), Expect = 1.7, Method: Composition-based stats.
Identities = 28/86 (33%), Positives = 40/86 (47%), Gaps = 22/86 (26%)
Query 57 SIPRVSKLQNFKDEYFEELVWMLPEIAESLKKKNNTDASGAFPQFKGLLKYVNIRDYQLF 116
S P S L + + FE LV M+ IA+S+ K+N+T SG +P + IR+Y F
Sbjct 428 SFPVASPLTTLQIQAFEGLVIMIHNIADSIDKENDTSPSGPYP--------IEIREYTPF 479
Query 117 AK--------------RLRKYLSKKI 128
+ RLRK +KI
Sbjct 480 WEDKPKDDSEAWVQFVRLRKAQKRKI 505
> clp:CPK_ORF00729 hypothetical protein
Length=121
Score = 36.6 bits (83), Expect = 2.4, Method: Compositional matrix adjust.
Identities = 33/90 (37%), Positives = 42/90 (47%), Gaps = 15/90 (17%)
Query 76 VWMLPEIAE-SLKKKNN----TDASGAFPQFKGLLKYVNIRDYQLFAKRLRKYLSKKIGK 130
VW + E SL +KN T PQ+ L+K QLF KRLR +S
Sbjct 37 VWSYRCVHEASLYEKNCFLTLTYDDKHLPQYGSLVKL----HLQLFLKRLRDRISP---- 88
Query 131 YEKIHSYVVSEYSPKTFRPHFHILFF-FDS 159
KI + EY K RPH+H+L F +DS
Sbjct 89 -HKIRYFGCGEYGTKLQRPHYHLLIFNYDS 117
> bfu:BC1G_05882 hypothetical protein
Length=506
Score = 38.5 bits (88), Expect = 3.0, Method: Compositional matrix adjust.
Identities = 28/91 (31%), Positives = 47/91 (52%), Gaps = 11/91 (12%)
Query 7 VNKNIPDFMSAKVNRPYMLEHLRLLEADRYKALALRYPNFISKFRPFILRSIPRVSKLQN 66
++ IP MS +PY + H L AD Y+ LA RYP+ K+ I R +
Sbjct 43 MSNKIP-LMSDVEEKPYCIWHPHLATADTYRELAQRYPDM--KYH------IGRACAVGG 93
Query 67 FKDEYFEELVWMLPEIAESLKKKNNTDASGA 97
+ D Y E + +LP+I+ + + +N+D +G+
Sbjct 94 YIDLYRE--LDLLPDISIAEEAWHNSDVTGS 122
> siy:YG5714_0844 hypothetical protein
Length=93
Score = 35.4 bits (80), Expect = 5.0, Method: Composition-based stats.
Identities = 21/54 (39%), Positives = 30/54 (56%), Gaps = 4/54 (7%)
Query 394 LKNQLSLQELVFAELGYSDELFDSFYVKPHIKVLKNAYIDKWKDVNYKEVHYFR 447
+ +L L EL AE+ YS+E VK + K +K +DKW+ VN+ YFR
Sbjct 40 IAEELKLNELK-AEITYSEEATTYVEVKLYKKAIKGTLLDKWRRVNFS---YFR 89
> caa:Caka_1841 TetR family transcriptional regulator
Length=193
Score = 36.6 bits (83), Expect = 6.7, Method: Compositional matrix adjust.
Identities = 20/78 (26%), Positives = 39/78 (50%), Gaps = 3/78 (4%)
Query 98 FPQFKGLLKYVNIRDYQLFAKRLRKYLSKKIGKYEKIHSYVVSEYSPKTFRPHFHILFF- 156
F +GL+ +N Q AKR+ + ++IH++ S +T P L F
Sbjct 52 FDNLEGLIVAINTHSAQQLAKRIGRIHESGHSAEQRIHAFCRSYLDFQTDEPELWKLLFA 111
Query 157 --FDSDEIAKNFRQAVYQ 172
++D+++++FR AV++
Sbjct 112 APINNDKLSQDFRDAVHK 129
Lambda K H a alpha
0.325 0.140 0.424 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 981216274224