bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-7_CDS_annotation_glimmer3.pl_2_6
Length=131
Score E
Sequences producing significant alignments: (Bits) Value
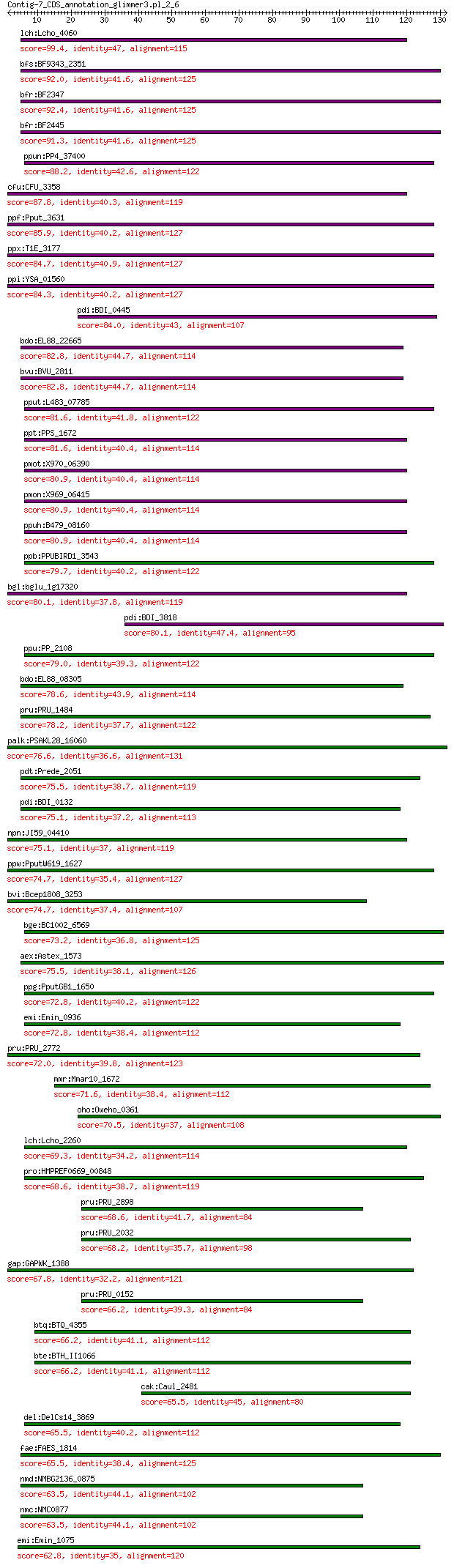
lch:Lcho_4060 peptidase M15A 99.4 3e-23
bfs:BF9343_2351 hypothetical protein 92.0 2e-21
bfr:BF2347 hypothetical protein 92.4 2e-21
bfr:BF2445 hypothetical protein 91.3 4e-21
ppun:PP4_37400 hypothetical protein 88.2 7e-20
cfu:CFU_3358 hypothetical protein 87.8 1e-19
ppf:Pput_3631 peptidase M15A 85.9 7e-19
ppx:T1E_3177 peptidase M15A 84.7 1e-18
ppi:YSA_01560 peptidase M15A 84.3 2e-18
pdi:BDI_0445 hypothetical protein 84.0 4e-18
bdo:EL88_22665 peptidase M15 82.8 8e-18
bvu:BVU_2811 hypothetical protein 82.8 8e-18
pput:L483_07785 peptidase M15 81.6 2e-17
ppt:PPS_1672 peptidase M15A 81.6 2e-17
pmot:X970_06390 peptidase M15 80.9 5e-17
pmon:X969_06415 peptidase M15 80.9 5e-17
ppuh:B479_08160 peptidase M15A 80.9 5e-17
ppb:PPUBIRD1_3543 peptidase M15A 79.7 1e-16
bgl:bglu_1g17320 peptidase M15A 80.1 1e-16
pdi:BDI_3818 hypothetical protein 80.1 1e-16
ppu:PP_2108 peptidase M15A 79.0 2e-16
bdo:EL88_08305 peptidase M15 78.6 2e-16
pru:PRU_1484 hypothetical protein 78.2 6e-16
palk:PSAKL28_16060 peptidase M15A 76.6 2e-15
pdt:Prede_2051 hypothetical protein 75.5 5e-15
pdi:BDI_0132 hypothetical protein 75.1 5e-15
npn:JI59_04410 hypothetical protein 75.1 7e-15
ppw:PputW619_1627 peptidase M15A 74.7 8e-15
bvi:Bcep1808_3253 peptidase M15A 74.7 8e-15
bge:BC1002_6569 peptidase M15A 73.2 4e-14
aex:Astex_1573 peptidase m15a 75.5 4e-14
ppg:PputGB1_1650 peptidase M15A 72.8 4e-14
emi:Emin_0936 peptidase M15A 72.8 5e-14
pru:PRU_2772 hypothetical protein 72.0 1e-13
mmr:Mmar10_1672 peptidase M15A 71.6 1e-13
oho:Oweho_0361 Peptidase M15 70.5 4e-13
lch:Lcho_2260 peptidase M15A 69.3 1e-12
pro:HMPREF0669_00848 hypothetical protein 68.6 2e-12
pru:PRU_2898 hypothetical protein 68.6 2e-12
pru:PRU_2032 hypothetical protein 68.2 3e-12
gap:GAPWK_1388 hypothetical protein 67.8 3e-12
pru:PRU_0152 hypothetical protein 66.2 1e-11
btq:BTQ_4355 peptidase M15 family protein 66.2 1e-11
bte:BTH_II1066 hypothetical protein 66.2 1e-11
cak:Caul_2481 peptidase M15A 65.5 2e-11
del:DelCs14_3869 peptidase M15A 65.5 2e-11
fae:FAES_1814 peptidase M15A 65.5 2e-11
nmd:NMBG2136_0875 Peptidase M15 family protein 63.5 1e-10
nmc:NMC0877 ATP binding protein 63.5 1e-10
emi:Emin_1075 peptidase M15A 62.8 2e-10
> lch:Lcho_4060 peptidase M15A
Length=236
Score = 99.4 bits (246), Expect = 3e-23, Method: Compositional matrix adjust.
Identities = 54/121 (45%), Positives = 72/121 (60%), Gaps = 6/121 (5%)
Query 5 KYFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
++F+ E+ + + E +N P +I L LC +LDP+RE GKPI VTSG+R P
Sbjct 5 RFFTLAELTHSNTAKAEGIDNQPTTTEIEALRALCTAVLDPLREAIGKPIKVTSGYRGPV 64
Query 63 LNRLVGGVSTSQHMQGLAADLRCDDPK--ALFDLIAESDLPFDQLIYY--QKKKFVHVSY 118
LNR V G + SQH++G AADL+ ALF + LPFDQLIY K+VHVS+
Sbjct 65 LNRRVKGAAKSQHLRGEAADLQSPGTAVLALFKRVIRLGLPFDQLIYEVNGASKWVHVSH 124
Query 119 S 119
S
Sbjct 125 S 125
> bfs:BF9343_2351 hypothetical protein
Length=131
Score = 92.0 bits (227), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 52/130 (40%), Positives = 76/130 (58%), Gaps = 6/130 (5%)
Query 5 KYFSFTEMIL--TASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
KYF+ E+ TA +N P + ++ L EL +LDP+RE YGKPI V+SG+R+
Sbjct 2 KYFTIKELSHSDTAVARGIDNTPTGEAVHNLTELVENVLDPLREKYGKPIRVSSGYRSAV 61
Query 63 LNRLVGGVSTSQHMQGLAADLRCDDPKA---LFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
LNR V G ++SQH G AAD+ + LF++I +LPFDQLI + +VHVS+
Sbjct 62 LNRSVNGATSSQHRLGQAADITVGSKEGNRRLFEII-RKELPFDQLIDEKDFSWVHVSFR 120
Query 120 PTYRHEVIVK 129
+ ++K
Sbjct 121 KGKNRKQVLK 130
> bfr:BF2347 hypothetical protein
Length=140
Score = 92.4 bits (228), Expect = 2e-21, Method: Compositional matrix adjust.
Identities = 52/130 (40%), Positives = 78/130 (60%), Gaps = 6/130 (5%)
Query 5 KYFSFTEMIL--TASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
KYF+ E+ TA +N P + ++ L EL +LDP+RE YGKPI V+SG+R+
Sbjct 11 KYFTIKELSHSDTAVARGIDNTPTGEVVHNLTELVENVLDPLREKYGKPIRVSSGYRSAV 70
Query 63 LNRLVGGVSTSQHMQGLAADLRCDDPKA---LFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
LNR V G ++SQH+ G AAD+ + LF++I + +LPFDQLI + +VHVS+
Sbjct 71 LNRSVNGATSSQHLLGQAADITVGSKEGNRRLFEIIRK-ELPFDQLIDEKDFSWVHVSFR 129
Query 120 PTYRHEVIVK 129
+ ++K
Sbjct 130 TGKNRKQVLK 139
> bfr:BF2445 hypothetical protein
Length=131
Score = 91.3 bits (225), Expect = 4e-21, Method: Compositional matrix adjust.
Identities = 52/130 (40%), Positives = 79/130 (61%), Gaps = 6/130 (5%)
Query 5 KYFSFTEMIL--TASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
KYF+ E+ TA +N P + I+ L +L +LDP+RE YGKPI V+SG+R+
Sbjct 2 KYFTIKELSHSDTAVARGIDNYPTAEAIHNLTKLVENVLDPLREKYGKPIRVSSGYRSAI 61
Query 63 LNRLVGGVSTSQHMQGLAADLRC---DDPKALFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
LNR V G ++SQH G AAD+ ++ + LF++I + +LPFDQLI + +VHVS+
Sbjct 62 LNRSVNGATSSQHRLGEAADITVGSKEENRKLFEIIRQ-ELPFDQLIDEKDFSWVHVSFC 120
Query 120 PTYRHEVIVK 129
+ ++K
Sbjct 121 EGRSRKQVLK 130
> ppun:PP4_37400 hypothetical protein
Length=143
Score = 88.2 bits (217), Expect = 7e-20, Method: Compositional matrix adjust.
Identities = 52/127 (41%), Positives = 81/127 (64%), Gaps = 7/127 (6%)
Query 6 YFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFL 63
+F+F EM ++ A++ +N P + L LC L+ VR ++G PI+V+SGFR+ +
Sbjct 6 HFTFDEMTVSQLAARDGFDNTPPPEARANLQLLC-GALEQVRALFGAPIIVSSGFRSEKV 64
Query 64 NRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHVSYSP- 120
NRL+GG + SQH+QGLAAD + P+ I+ES +PFDQLI + ++VH+S +P
Sbjct 65 NRLIGGATNSQHVQGLAADFTVIEVSPRETVRRISESAVPFDQLI-LEFDRWVHLSVAPG 123
Query 121 TYRHEVI 127
T R +V+
Sbjct 124 TPRRQVL 130
> cfu:CFU_3358 hypothetical protein
Length=143
Score = 87.8 bits (216), Expect = 1e-19, Method: Compositional matrix adjust.
Identities = 48/123 (39%), Positives = 76/123 (62%), Gaps = 6/123 (5%)
Query 1 MYRPKYFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGF 58
M ++F+ EM ++ + ++ +N P I+ L +L L+ +R + GKPI+VTSG+
Sbjct 1 MNLTEHFTLAEMTISETAVRQGIDNTPSLAVIDNL-KLTAATLEQIRTLVGKPIIVTSGY 59
Query 59 RTPFLNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHV 116
R+P +N+ VGG S S H+ GLAAD+ PKAL +LI +S + FDQLI + ++VHV
Sbjct 60 RSPAVNKAVGGASNSAHVLGLAADINVPGYTPKALANLIKDSGIQFDQLI-LEFDRWVHV 118
Query 117 SYS 119
+
Sbjct 119 GLA 121
> ppf:Pput_3631 peptidase M15A
Length=143
Score = 85.9 bits (211), Expect = 7e-19, Method: Compositional matrix adjust.
Identities = 51/132 (39%), Positives = 82/132 (62%), Gaps = 7/132 (5%)
Query 1 MYRPKYFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGF 58
M+ +F+F EM ++ A+E +N P + L LC + L+ VR ++ PI+++SG+
Sbjct 1 MFITPHFTFDEMTVSQLAAREGFDNAPPLEARANLQLLC-YALEQVRALFDAPIIISSGY 59
Query 59 RTPFLNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHV 116
R+ +NRL+GG S SQH+QGLAAD + P+ ++ES +PFDQLI + K+VH+
Sbjct 60 RSERVNRLIGGASDSQHVQGLAADFTVIEVSPRETVRRVSESTVPFDQLI-LEFDKWVHL 118
Query 117 SYS-PTYRHEVI 127
S + T R +V+
Sbjct 119 SVTRGTPRRQVL 130
> ppx:T1E_3177 peptidase M15A
Length=143
Score = 84.7 bits (208), Expect = 1e-18, Method: Compositional matrix adjust.
Identities = 52/132 (39%), Positives = 81/132 (61%), Gaps = 7/132 (5%)
Query 1 MYRPKYFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGF 58
M+ +F+F EM ++ A+E +N P + L LC L+ VR ++ PI+++SG+
Sbjct 1 MFITPHFTFDEMTVSQLAAREGFDNAPPLEARANLQLLC-CALEQVRALFDAPIIISSGY 59
Query 59 RTPFLNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHV 116
R+ +NRL+GG S SQH+QGLAAD + P+ I+ES +PFDQLI + K+VH+
Sbjct 60 RSERVNRLIGGASDSQHVQGLAADFTVIEISPRETVRRISESAVPFDQLI-LEFDKWVHL 118
Query 117 SYS-PTYRHEVI 127
S + T R +V+
Sbjct 119 SVTRGTPRRQVL 130
> ppi:YSA_01560 peptidase M15A
Length=143
Score = 84.3 bits (207), Expect = 2e-18, Method: Compositional matrix adjust.
Identities = 51/132 (39%), Positives = 81/132 (61%), Gaps = 7/132 (5%)
Query 1 MYRPKYFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGF 58
M+ +F+F EM ++ A+E +N P + L LC L+ VR ++ PI+++SG+
Sbjct 1 MFITPHFTFDEMTVSQLAAREGFDNAPPLEARANLQLLC-CALEQVRALFDAPIIISSGY 59
Query 59 RTPFLNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHV 116
R+ +NRL+GG S SQH+QGLAAD + P+ ++ES +PFDQLI + K+VH+
Sbjct 60 RSERVNRLIGGASDSQHVQGLAADFTVIEVSPRETVRRVSESTVPFDQLI-LEFDKWVHL 118
Query 117 SYS-PTYRHEVI 127
S + T R +V+
Sbjct 119 SVTRGTPRRQVL 130
> pdi:BDI_0445 hypothetical protein
Length=156
Score = 84.0 bits (206), Expect = 4e-18, Method: Compositional matrix adjust.
Identities = 46/110 (42%), Positives = 66/110 (60%), Gaps = 3/110 (3%)
Query 22 NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAA 81
NN P L L +L+P+R +YG PI + SG+R +NRL GGV TSQH++G AA
Sbjct 29 NNQPPPCACLALQNLAVGLLEPLRLLYGAPIAILSGYRNEKVNRLAGGVVTSQHLKGEAA 88
Query 82 DLR-CDDPKALFDLIAESDLPFDQLIYYQKKKFVHVSY--SPTYRHEVIV 128
D D P+ L D++ S L FDQ I Y +++F+H+S + R +V+V
Sbjct 89 DCYVADGPEKLLDVLQCSGLVFDQAILYGRRRFLHLSLKINGNNRMQVLV 138
> bdo:EL88_22665 peptidase M15
Length=132
Score = 82.8 bits (203), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 51/119 (43%), Positives = 66/119 (55%), Gaps = 6/119 (5%)
Query 5 KYFSFTEMI--LTASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
K+F+ E+ TA NN + + L L +LDP+R +GKPI V SG+R P
Sbjct 2 KFFTIAELCKSTTADRLGINNRCRQEHVTALTALVDNVLDPLRTWWGKPITVNSGYRCPE 61
Query 63 LNRLVGGVSTSQHMQGLAADLRCDD---PKALFDLIAESDLPFDQLIYYQKKKFVHVSY 118
LN V G TSQHM+G AAD+ D K LF+ I +LP+DQLI +VHVSY
Sbjct 62 LNAAVKGSKTSQHMKGEAADIDTGDRQQNKLLFEYI-RKNLPYDQLIDESNFAWVHVSY 119
> bvu:BVU_2811 hypothetical protein
Length=132
Score = 82.8 bits (203), Expect = 8e-18, Method: Compositional matrix adjust.
Identities = 51/119 (43%), Positives = 66/119 (55%), Gaps = 6/119 (5%)
Query 5 KYFSFTEMI--LTASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
K+F+ E+ TA NN + + L L +LDP+R +GKPI V SG+R P
Sbjct 2 KFFTIAELCKSTTADRLGINNRCRQEHVTALTALVDNVLDPLRTWWGKPITVNSGYRCPE 61
Query 63 LNRLVGGVSTSQHMQGLAADLRCDD---PKALFDLIAESDLPFDQLIYYQKKKFVHVSY 118
LN V G TSQHM+G AAD+ D K LF+ I +LP+DQLI +VHVSY
Sbjct 62 LNAAVKGSKTSQHMKGEAADIDTGDRQQNKLLFEYI-RKNLPYDQLIDESNFAWVHVSY 119
> pput:L483_07785 peptidase M15
Length=143
Score = 81.6 bits (200), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 51/127 (40%), Positives = 78/127 (61%), Gaps = 7/127 (6%)
Query 6 YFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFL 63
+F+ EM ++ A++ +N P D L LC L+ VR ++G PI+V+SGFR+ +
Sbjct 6 HFTLDEMTVSQLAARDGFDNTPPPDARANLQLLC-CALEQVRALFGVPIIVSSGFRSEKV 64
Query 64 NRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHVSYS-P 120
NRL+GG + SQH+QGLAAD + P+ I ES +PFDQLI + ++VH+S +
Sbjct 65 NRLIGGATNSQHIQGLAADFTVMEVSPRETARRINESAVPFDQLI-LEFDRWVHLSVTRG 123
Query 121 TYRHEVI 127
T R +V+
Sbjct 124 TPRRQVL 130
> ppt:PPS_1672 peptidase M15A
Length=143
Score = 81.6 bits (200), Expect = 2e-17, Method: Compositional matrix adjust.
Identities = 46/119 (39%), Positives = 74/119 (62%), Gaps = 8/119 (7%)
Query 6 YFSFTEMILTASTAQE---NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
+F+ EM ++ A+E NN P N L+L L+ VR ++G P++V+SG+R+P
Sbjct 6 HFTLDEMTVSQLAAREGLDNNPPAEARAN--LQLLCNALEQVRALFGAPVIVSSGYRSPA 63
Query 63 LNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
+N+ +GG TS+H+QGLAAD D P+ + ++ES +PFDQLI + +VH++ S
Sbjct 64 VNQRIGGTLTSKHLQGLAADFTVIDVSPREVVRRVSESTIPFDQLI-LEFDDWVHLAVS 121
> pmot:X970_06390 peptidase M15
Length=143
Score = 80.9 bits (198), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 46/119 (39%), Positives = 74/119 (62%), Gaps = 8/119 (7%)
Query 6 YFSFTEMILTASTAQE---NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
+F+ EM ++ A+E NN P N L+L L+ VR ++G P++V+SG+R+P
Sbjct 6 HFTLDEMTVSQLAAREGLDNNPPAEARAN--LQLLCNALEQVRALFGAPVIVSSGYRSPA 63
Query 63 LNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
+N+ +GG TS+H+QGLAAD + P+ + I+ES +PFDQLI + +VH++ S
Sbjct 64 VNQRIGGTLTSKHLQGLAADFTVIEVTPREVVRRISESTIPFDQLI-LEFDDWVHLAVS 121
> pmon:X969_06415 peptidase M15
Length=143
Score = 80.9 bits (198), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 46/119 (39%), Positives = 74/119 (62%), Gaps = 8/119 (7%)
Query 6 YFSFTEMILTASTAQE---NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
+F+ EM ++ A+E NN P N L+L L+ VR ++G P++V+SG+R+P
Sbjct 6 HFTLDEMTVSQLAAREGLDNNPPAEARAN--LQLLCNALEQVRALFGAPVIVSSGYRSPA 63
Query 63 LNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
+N+ +GG TS+H+QGLAAD + P+ + I+ES +PFDQLI + +VH++ S
Sbjct 64 VNQRIGGTLTSKHLQGLAADFTVIEVTPREVVRRISESTIPFDQLI-LEFDDWVHLAVS 121
> ppuh:B479_08160 peptidase M15A
Length=143
Score = 80.9 bits (198), Expect = 5e-17, Method: Compositional matrix adjust.
Identities = 46/119 (39%), Positives = 74/119 (62%), Gaps = 8/119 (7%)
Query 6 YFSFTEMILTASTAQE---NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
+F+ EM ++ A+E NN P N L+L L+ VR ++G P++V+SG+R+P
Sbjct 6 HFTLDEMTVSQLAAREGLDNNPPAEARAN--LQLLCNALEQVRALFGAPVIVSSGYRSPA 63
Query 63 LNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
+N+ +GG TS+H+QGLAAD + P+ + I+ES +PFDQLI + +VH++ S
Sbjct 64 VNQRIGGTLTSKHLQGLAADFTVIEVTPREVVRRISESTIPFDQLI-LEFDDWVHLAVS 121
> ppb:PPUBIRD1_3543 peptidase M15A
Length=143
Score = 79.7 bits (195), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 49/127 (39%), Positives = 78/127 (61%), Gaps = 7/127 (6%)
Query 6 YFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFL 63
+F+ EM ++ A++ +N P + L LC L+ VR ++ PI+++SG+R+ +
Sbjct 6 HFTLDEMTVSQLAARDGFDNAPPHEARANLQLLC-GALEQVRALFDAPIIISSGYRSERV 64
Query 64 NRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHVSYS-P 120
NRL+GG S SQH+QGLAAD + P+ I+ES +PFDQLI + K+VH+S +
Sbjct 65 NRLIGGASDSQHVQGLAADFTVIEVSPRETVRRISESAVPFDQLI-LEFDKWVHLSVTRG 123
Query 121 TYRHEVI 127
T R +V+
Sbjct 124 TPRRQVL 130
> bgl:bglu_1g17320 peptidase M15A
Length=149
Score = 80.1 bits (196), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 45/124 (36%), Positives = 70/124 (56%), Gaps = 6/124 (5%)
Query 1 MYRPKYFSFTEMILT--ASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGF 58
M +F+ E+ + A T Q +N P + L L + + + GKP+ +TSG+
Sbjct 1 MQLTDHFTLEELTASDVARTRQIDNTPSAATVENLRRLAQTLEQARVLLGGKPMQITSGY 60
Query 59 RTPFLNRLVGGVSTSQHMQGLAADLRCDDPKALFDLI---AESDLPFDQLIYYQKKKFVH 115
R+P LNR VGGV++S H+ GLAAD C A D++ A S+L FDQLI ++ ++VH
Sbjct 61 RSPALNRAVGGVASSAHLAGLAADFVCPKFGAPLDVVRKLAASNLAFDQLI-HEGGRWVH 119
Query 116 VSYS 119
+ +
Sbjct 120 IGLT 123
> pdi:BDI_3818 hypothetical protein
Length=167
Score = 80.1 bits (196), Expect = 1e-16, Method: Compositional matrix adjust.
Identities = 45/103 (44%), Positives = 63/103 (61%), Gaps = 8/103 (8%)
Query 36 LCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAADLRC---DDPKALF 92
L +LDP+R ++G+PIV++SG+R P LN L+GG S H+ G AADL DD + LF
Sbjct 61 LVEMLLDPLRRVWGRPIVISSGYRCPELNILIGGAKHSHHLLGCAADLIAGSPDDHRLLF 120
Query 93 DLIAES----DLPFDQLIYYQKKKFVHVSYSP-TYRHEVIVKD 130
LI E+ L F QLI +++H+SY P R +VI K+
Sbjct 121 RLIQETHELCGLEFTQLILEPGARWIHISYVPGNLRCQVIDKE 163
> ppu:PP_2108 peptidase M15A
Length=143
Score = 79.0 bits (193), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 48/127 (38%), Positives = 78/127 (61%), Gaps = 7/127 (6%)
Query 6 YFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFL 63
+F+ EM ++ A+E +N P + L LC L+ VR ++ PI+++SG+R+ +
Sbjct 6 HFTLDEMTVSQLAAREGFDNAPPLEARANLQLLC-GALEQVRALFDAPIIISSGYRSERV 64
Query 64 NRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHVSYS-P 120
NRL+GG S SQH+QGLAAD + P+ I++S +PFDQLI + ++VH+S +
Sbjct 65 NRLIGGASDSQHVQGLAADFTVIEVSPRETVRRISKSAVPFDQLI-LEFDRWVHLSVTRG 123
Query 121 TYRHEVI 127
T R +V+
Sbjct 124 TPRRQVL 130
> bdo:EL88_08305 peptidase M15
Length=132
Score = 78.6 bits (192), Expect = 2e-16, Method: Compositional matrix adjust.
Identities = 50/119 (42%), Positives = 65/119 (55%), Gaps = 6/119 (5%)
Query 5 KYFSFTEMI--LTASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
K+F+ E+ TA NN + + L L +LDP+R +GKPI V SG+R
Sbjct 2 KFFTIAELCKSTTADRLGINNRCRQEHVTALTALVDNVLDPLRTWWGKPITVNSGYRCLE 61
Query 63 LNRLVGGVSTSQHMQGLAADLRCDD---PKALFDLIAESDLPFDQLIYYQKKKFVHVSY 118
LN V G TSQHM+G AAD+ D K LF+ I +LP+DQLI +VHVSY
Sbjct 62 LNAAVKGSKTSQHMKGEAADIDTGDRQQNKLLFEYI-RKNLPYDQLIDESNFAWVHVSY 119
> pru:PRU_1484 hypothetical protein
Length=155
Score = 78.2 bits (191), Expect = 6e-16, Method: Compositional matrix adjust.
Identities = 46/137 (34%), Positives = 72/137 (53%), Gaps = 16/137 (12%)
Query 5 KYFSFTEMILTASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFLN 64
++F+ EM ++S + N+P + I L LC W L+ +RE G+ IV+ SG+R+P LN
Sbjct 15 EHFTLGEMTKSSSHPEVYNIPSHEAIANLKRLCTW-LEALRERTGRSIVINSGYRSPQLN 73
Query 65 RLVGGVSTSQHMQGLAADLRCDDPKALFDLIA-------ESDLPFDQLIYYQKK---KFV 114
R VGG TS H+ G AAD+R + A ES FD+L+ + + ++
Sbjct 74 RKVGGAPTSNHLTGCAADIRTSGMEQAITYAAILLAYARESQQEFDELLIEKNRYGAVWL 133
Query 115 HVSYSPTY-----RHEV 126
H + P+ RH+V
Sbjct 134 HFAVRPSTPSSKNRHKV 150
> palk:PSAKL28_16060 peptidase M15A
Length=143
Score = 76.6 bits (187), Expect = 2e-15, Method: Compositional matrix adjust.
Identities = 48/137 (35%), Positives = 78/137 (57%), Gaps = 8/137 (6%)
Query 1 MYRPKYFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGF 58
MY ++F+ EM ++ + A+ +N PD + L LC + L+ +R++ KP++V+SG+
Sbjct 1 MYLSEHFTLAEMTVSETAARLGIDNTPDAQTLVNLRRLCTF-LEQIRQLVDKPVLVSSGY 59
Query 59 RTPFLNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHV 116
R LN ++GG S H GLAAD+ P+ L +A+S L FDQLI + ++VHV
Sbjct 60 RCLQLNEVIGGSLQSAHTDGLAADINVPGLTPRELARGVADSSLMFDQLI-LEFDQWVHV 118
Query 117 --SYSPTYRHEVIVKDN 131
S +P R + V+
Sbjct 119 GLSLTPERRQLLTVRKG 135
> pdt:Prede_2051 hypothetical protein
Length=142
Score = 75.5 bits (184), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 46/127 (36%), Positives = 72/127 (57%), Gaps = 9/127 (7%)
Query 5 KYFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
++F++ E+ + + + +N P+ L L +L+P+R +G PIVV+SGFR+P
Sbjct 11 EHFTWYELTRSGAALEHALDNTPNDRQRQALRALAENVLEPLRRRFG-PIVVSSGFRSPA 69
Query 63 LNRLVGGVSTSQHMQGLAADLRCDDPKALFDLIAE--SDLPFDQLIY----YQKKKFVHV 116
+NRLVGGV SQH++G AAD+ D + L FDQLI+ +++HV
Sbjct 70 VNRLVGGVPGSQHLRGEAADIVIGDVDRGLHICHHIRRHLDFDQLIFEPLGSPTPRWLHV 129
Query 117 SYSPTYR 123
SY+ T R
Sbjct 130 SYTATRR 136
> pdi:BDI_0132 hypothetical protein
Length=142
Score = 75.1 bits (183), Expect = 5e-15, Method: Compositional matrix adjust.
Identities = 42/116 (36%), Positives = 66/116 (57%), Gaps = 4/116 (3%)
Query 5 KYFSFTEMILTASTAQ---ENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTP 61
++F E + +A + N LP + I + L +L+P+R + +P+ + SG+R+
Sbjct 10 EHFRMDEFVYSAMAVELGLNNALPS-EVIPAIRNLMVRLLEPLRIYHRQPMYIMSGYRSE 68
Query 62 FLNRLVGGVSTSQHMQGLAADLRCDDPKALFDLIAESDLPFDQLIYYQKKKFVHVS 117
LNRLVGG +SQHM+G A D+ D L + + S L FDQ I Y+ K F+H+S
Sbjct 69 ELNRLVGGAPSSQHMKGEAVDIYTVDRNRLLEDLVASRLNFDQAILYRTKGFIHLS 124
> npn:JI59_04410 hypothetical protein
Length=149
Score = 75.1 bits (183), Expect = 7e-15, Method: Compositional matrix adjust.
Identities = 44/124 (35%), Positives = 68/124 (55%), Gaps = 6/124 (5%)
Query 1 MYRPKYFSFTEMI--LTASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGF 58
M K+F+ EM+ TA +N P + + L LC +L+PVR + P++V+SG+
Sbjct 1 MQLSKHFTLEEMVRSQTAERLGIDNTPSQEIVANLRGLCAHVLEPVRAHFNSPVIVSSGY 60
Query 59 RTPFLNRLVGGVSTSQHMQGLAADLRC-DDPK-ALFDLIAESDLPFDQLIY-YQKKKFVH 115
R P LN +GG +SQH +G A D P +F I +L +DQLIY + + ++H
Sbjct 61 RCPQLNVAIGGSKSSQHCKGEAGDFSVLGQPNITVFKWIWH-NLDYDQLIYEFGESGWIH 119
Query 116 VSYS 119
S+S
Sbjct 120 ASFS 123
> ppw:PputW619_1627 peptidase M15A
Length=143
Score = 74.7 bits (182), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 45/132 (34%), Positives = 77/132 (58%), Gaps = 7/132 (5%)
Query 1 MYRPKYFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGF 58
M+ F+ EMI++ ++E +N P I+ L LC L+ VR ++G P++++SG+
Sbjct 1 MFITPNFTLEEMIVSQVASREGLDNTPSQQVISNLHLLCDA-LEQVRALFGLPVIISSGY 59
Query 59 RTPFLNRLVGGVSTSQHMQGLAADLRC--DDPKALFDLIAESDLPFDQLIYYQKKKFVHV 116
R+P LN+ +GG SQH++GLAAD + + ++ S + FDQ+I + +VH+
Sbjct 60 RSPALNKRIGGSPRSQHLRGLAADFEIFGISNREVVRQVSASAVSFDQII-LEFDSWVHL 118
Query 117 SYSPTY-RHEVI 127
S +P R EV+
Sbjct 119 SVTPGVPRREVL 130
> bvi:Bcep1808_3253 peptidase M15A
Length=149
Score = 74.7 bits (182), Expect = 8e-15, Method: Compositional matrix adjust.
Identities = 40/111 (36%), Positives = 61/111 (55%), Gaps = 4/111 (4%)
Query 1 MYRPKYFSFTEMILTASTAQEN--NLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGF 58
M ++F E +++ + A+ N P + I L LC+ +L P+R +P+V+TSG+
Sbjct 1 MQLSEHFELAEFLVSETAARRGIANEPTPEVIENLRRLCQSVLQPLRVHLKRPVVITSGY 60
Query 59 RTPFLNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIY 107
R+P LNR +GG TS HMQG AADL P + + LP Q+I+
Sbjct 61 RSPALNRAIGGSPTSHHMQGRAADLIVPGMTPLLVCQAAHQLKLPCVQIIH 111
> bge:BC1002_6569 peptidase M15A
Length=150
Score = 73.2 bits (178), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 46/134 (34%), Positives = 73/134 (54%), Gaps = 12/134 (9%)
Query 6 YFSFTEMIL--TASTAQENNLPDWDDINRLLELCRWILDPVREMY-GKPIVVTSGFRTPF 62
+F+ E+ TA+ + +N P + L + L+ VR + +P++V+SG+R+P
Sbjct 7 HFTLEELTQSETATRRRIDNTPSPAIVENLTRTAQ-TLEQVRALLCSRPVLVSSGYRSPA 65
Query 63 LNRLVGGVSTSQHMQGLAADLRC---DDPKALFDLIAESDLPFDQLIYYQKKKFVHVSYS 119
LN VGG + S HM GLAAD C P + IA S++PFDQLI Q+ +VH+ +
Sbjct 66 LNTAVGGAANSAHMTGLAADFICPGFGSPLEICRKIAASNIPFDQLI--QEGTWVHIGLA 123
Query 120 PT---YRHEVIVKD 130
P R +V+ +
Sbjct 124 PGGQKARQQVLTAN 137
> aex:Astex_1573 peptidase m15a
Length=361
Score = 75.5 bits (184), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 48/132 (36%), Positives = 71/132 (54%), Gaps = 10/132 (8%)
Query 5 KYFSFTEMILTASTAQEN---NLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTP 61
+YF+ E+ + A+ N N+P + +N L + + +D VR + G+PI V SG+R P
Sbjct 221 QYFTLEELT-QSDIAERNGIENMPTPEHLNNLKDTAQR-MDKVRALLGQPITVRSGYRGP 278
Query 62 FLNRLVGGVSTSQHMQGLAADL---RCDDPKALFDLIAESDLPFDQLIYYQKKKFVHVSY 118
LN +GG TS HM G A D R P + I SD+ FDQLIY + +VH+ +
Sbjct 279 ALNAKIGGSKTSAHMIGRAVDFVSQRFGTPLDICRKIMASDIVFDQLIY--EGTWVHIGF 336
Query 119 SPTYRHEVIVKD 130
S T R + + D
Sbjct 337 SDTPRRQALRAD 348
> ppg:PputGB1_1650 peptidase M15A
Length=143
Score = 72.8 bits (177), Expect = 4e-14, Method: Compositional matrix adjust.
Identities = 49/127 (39%), Positives = 78/127 (61%), Gaps = 7/127 (6%)
Query 6 YFSFTEMILTASTAQE--NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFL 63
+F+F EM ++ A++ +N P + L L L+ VR ++ PI+V+SG+R+ +
Sbjct 6 HFTFDEMTVSQLAARDGFDNTPP-PEARANLLLLCCALEQVRALFDAPIIVSSGYRSEKV 64
Query 64 NRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIYYQKKKFVHVSYS-P 120
NRL+GG +SQH+QGLAAD + P+ I+ES +PFDQLI + K+VH+S +
Sbjct 65 NRLIGGAVSSQHVQGLAADFTVVEVSPRETVRRISESGVPFDQLI-LEFDKWVHLSVARD 123
Query 121 TYRHEVI 127
T R +V+
Sbjct 124 TPRRQVL 130
> emi:Emin_0936 peptidase M15A
Length=151
Score = 72.8 bits (177), Expect = 5e-14, Method: Compositional matrix adjust.
Identities = 43/120 (36%), Positives = 73/120 (61%), Gaps = 11/120 (9%)
Query 6 YFSFTEMILTASTAQENNLPDWD----DINRLLELCRWILDPVREMYGKPIVVTSGFRTP 61
+FSF E++ T++ ++ + +W+ ++++L++L + + VR + G P+++TS R P
Sbjct 15 HFSFKELVNTSN--EKFVMANWNYGLLNMSQLVKLANF-GETVRTVLGVPMIITSAIRCP 71
Query 62 FLNRLVGGVSTSQHMQGLAADLRCDDPKA--LFDLIAESDLPFDQLIYYQK--KKFVHVS 117
LN +GG TSQHM+ A D C +FDLI ES+L F QLI Q K+++H+S
Sbjct 72 ELNESIGGSKTSQHMKCEAIDFICRGIGVARIFDLIRESNLTFGQLILEQAGGKEWIHIS 131
> pru:PRU_2772 hypothetical protein
Length=156
Score = 72.0 bits (175), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 49/139 (35%), Positives = 70/139 (50%), Gaps = 16/139 (12%)
Query 1 MYRPKYFSFTEMILTASTAQEN--NLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGF 58
M ++F+ EM +A+ + N+P I + LC L+PVR G PI V SG+
Sbjct 9 MRLSEHFTLGEMCYSATAEAKKIPNIPLKQHITAMQNLCERCLEPVRCQLGLPIKVNSGY 68
Query 59 RTPFLNRLVGGVSTSQHMQGLAADLRC----------DDPKALFDLI--AESDLPFDQLI 106
R LN++VGGV TSQH++G AAD+ D +A L+ AE FDQLI
Sbjct 69 RCQLLNQMVGGVPTSQHLKGEAADITIPRSHRPFGHPTDEQAARLLLKYAEQYADFDQLI 128
Query 107 YYQKKK--FVHVSYSPTYR 123
+ +VH+S +R
Sbjct 129 LEHRGNSWWVHISCRIDFR 147
> mmr:Mmar10_1672 peptidase M15A
Length=146
Score = 71.6 bits (174), Expect = 1e-13, Method: Compositional matrix adjust.
Identities = 43/115 (37%), Positives = 62/115 (54%), Gaps = 4/115 (3%)
Query 15 TASTAQENNLPDWDDINRLLELCRWILDPVREMYG-KPIVVTSGFRTPFLNRLVGGVSTS 73
TA + N PD + R L ++ VR + G + I +TSG+R P +N+ VGGVS S
Sbjct 16 TADELGDPNTPDEPHM-RALHGLACGMEQVRRILGDRAITITSGYRNPVVNKAVGGVSNS 74
Query 74 QHMQGLAAD--LRCDDPKALFDLIAESDLPFDQLIYYQKKKFVHVSYSPTYRHEV 126
H G AAD ++ +P + + S L FDQLIY ++ H+S+ P R EV
Sbjct 75 AHALGYAADFSVKGMEPVDVARALEASPLAFDQLIYEASRRINHISFDPRMRREV 129
> oho:Oweho_0361 Peptidase M15
Length=152
Score = 70.5 bits (171), Expect = 4e-13, Method: Compositional matrix adjust.
Identities = 40/117 (34%), Positives = 60/117 (51%), Gaps = 10/117 (9%)
Query 22 NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAA 81
+N P+ + L + I P+RE + PI ++SGFR LN +GG TS H +G A
Sbjct 24 DNTPNAQQLENLKRVAENIFQPIREHFNTPIFISSGFRCEDLNTRIGGSRTSSHCKGEAL 83
Query 82 DLRCDD-----PKALFDLIAESDLPFDQLIYY----QKKKFVHVSYSPTYRHEVIVK 129
D+ D + +FD + ++L FDQLIY Q +VH+SY T ++K
Sbjct 84 DIDMDGRGTITNRQVFDYV-RANLQFDQLIYEFGDDQNPGWVHISYKKTGNRNQVLK 139
> lch:Lcho_2260 peptidase M15A
Length=162
Score = 69.3 bits (168), Expect = 1e-12, Method: Compositional matrix adjust.
Identities = 39/122 (32%), Positives = 67/122 (55%), Gaps = 9/122 (7%)
Query 6 YFSFTEMILTASTAQEN--NLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFL 63
+F +E++++ S + N P + L +W L+ +R G PI V SG+R+P +
Sbjct 14 HFRLSELVISPSGERAGLRNEPQATQVRNLRRTAQW-LETLRGALGCPITVLSGYRSPAV 72
Query 64 NRLVGGVSTSQHMQGLAADLRC---DDPKALFDLIAESDLPFDQLIYYQ---KKKFVHVS 117
N+LVGG + S H++GLAAD P+A+ + + L FD+L++ Q + +VHV
Sbjct 73 NKLVGGSNQSAHLRGLAADFIAPAFGSPRAVCERVLSLGLSFDKLMFDQLIYEGSWVHVG 132
Query 118 YS 119
+
Sbjct 133 LA 134
> pro:HMPREF0669_00848 hypothetical protein
Length=158
Score = 68.6 bits (166), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 46/128 (36%), Positives = 69/128 (54%), Gaps = 11/128 (9%)
Query 6 YFSFTEMILTASTAQEN--NLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFL 63
+FS EM +A+ Q + N P+ I RL LC +L+P+R +G I +TSGFR+ L
Sbjct 17 HFSLKEMTSSATATQLHITNCPNATQIARLKALCEHVLEPLRNRFG-AIRITSGFRSERL 75
Query 64 NRLVGGVSTSQHMQGLAADLRCDDPK---ALFDLIAESDLPFDQLIYYQKKK----FVHV 116
N + S SQH G AAD+ + + +F I + FDQL+ +K+K ++HV
Sbjct 76 NSALCANSLSQHTFGEAADIYVPNRERGLEMFHFICQ-HCTFDQLLLERKRKTPSFWLHV 134
Query 117 SYSPTYRH 124
SY R+
Sbjct 135 SYKSDRRY 142
> pru:PRU_2898 hypothetical protein
Length=160
Score = 68.6 bits (166), Expect = 2e-12, Method: Compositional matrix adjust.
Identities = 35/91 (38%), Positives = 49/91 (54%), Gaps = 8/91 (9%)
Query 23 NLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAAD 82
N+P + I L LC W L+ +RE +PI++ SG+R+P LNR VGG TS H+ G A D
Sbjct 32 NIPSHEVIAHLKNLCTW-LEVLREKASQPIIINSGYRSPQLNRKVGGAPTSNHLTGCAVD 90
Query 83 LRCDD-------PKALFDLIAESDLPFDQLI 106
+R L + ES FD+L+
Sbjct 91 IRTSGYEQAIVYAAILINYAKESQQEFDELL 121
> pru:PRU_2032 hypothetical protein
Length=149
Score = 68.2 bits (165), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 35/108 (32%), Positives = 60/108 (56%), Gaps = 11/108 (10%)
Query 23 NLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAAD 82
N+P + I L +LC+W L+ +RE +PI++ SG+R+P LNR VGG + S H+ G A D
Sbjct 31 NIPSHEAIANLTKLCQW-LEFLRERSSQPIIINSGYRSPQLNRKVGGAANSNHLTGCAVD 89
Query 83 LRCDD-------PKALFDLIAESDLPFDQLIYYQKK---KFVHVSYSP 120
+R L D +++ FD+L+ + + ++H++ P
Sbjct 90 IRTSGYEQAIQYAAILIDYANKNNQQFDELLIERNRYGAVWLHLAVRP 137
> gap:GAPWK_1388 hypothetical protein
Length=146
Score = 67.8 bits (164), Expect = 3e-12, Method: Compositional matrix adjust.
Identities = 39/126 (31%), Positives = 68/126 (54%), Gaps = 8/126 (6%)
Query 1 MYRPKYFSFTEMILTASTAQ---ENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSG 57
M ++F+ E + + + +N++PD D+ + L L+ VRE PI+++SG
Sbjct 1 MKLTEHFTLEEFTRSTTATRLKIDNHVPD--DLMVNIHLTANKLESVREALTNPIIISSG 58
Query 58 FRTPFLNRLVGGVSTSQHMQGLAADLRC--DDPKALFDLIAESDLPFDQLIYYQKKKFVH 115
+R P LN VGG S H +GLA D C +PK + + + + FD++I + ++VH
Sbjct 59 YRCPALNNKVGGSQNSAHTKGLAVDFHCAYGNPKQICQRLIMAGVEFDKII-QEYNQWVH 117
Query 116 VSYSPT 121
+ +S T
Sbjct 118 IEFSHT 123
> pru:PRU_0152 hypothetical protein
Length=150
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 33/91 (36%), Positives = 48/91 (53%), Gaps = 8/91 (9%)
Query 23 NLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAAD 82
N+P + I L LC W L+ +RE PI++ SG+R+P NR VGG TS H+ G A D
Sbjct 32 NIPSHEAIANLKRLCEW-LEVLREKASHPIIINSGYRSPQFNRKVGGAPTSNHITGCAVD 90
Query 83 LRCDD-------PKALFDLIAESDLPFDQLI 106
+R + D ES+ +D+L+
Sbjct 91 IRTSGYEQAICYAAIIIDYAKESNQDYDELL 121
> btq:BTQ_4355 peptidase M15 family protein
Length=149
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 46/119 (39%), Positives = 65/119 (55%), Gaps = 9/119 (8%)
Query 9 FTEMILTAS-TAQENNLPDWDDINRLLELCRW--ILDPVRE-MYGKPIVVTSGFRTPFLN 64
FT LTAS TA+ + + L R +L+ VR+ + G+P+ +TSG+R LN
Sbjct 8 FTLEELTASDTARRRGIDNTPSAAVTANLRRTAEMLERVRDVLGGRPVRITSGYRAAALN 67
Query 65 RLVGGVSTSQHMQGLAADLRCDDPKALFDL---IAESDLPFDQLIYYQKKKFVHVSYSP 120
R VGGV +S H+ GLAAD C D+ I S + FDQLI Q+ +VH+ +P
Sbjct 68 RAVGGVPSSAHLSGLAADFVCPKIGTPLDICRAIRASPIEFDQLI--QEGTWVHIGLAP 124
> bte:BTH_II1066 hypothetical protein
Length=149
Score = 66.2 bits (160), Expect = 1e-11, Method: Compositional matrix adjust.
Identities = 46/119 (39%), Positives = 65/119 (55%), Gaps = 9/119 (8%)
Query 9 FTEMILTAS-TAQENNLPDWDDINRLLELCRW--ILDPVRE-MYGKPIVVTSGFRTPFLN 64
FT LTAS TA+ + + L R +L+ VR+ + G+P+ +TSG+R LN
Sbjct 8 FTLEELTASDTARRRGIDNTPSAAVTANLRRTAEMLERVRDVLGGRPVRITSGYRAAALN 67
Query 65 RLVGGVSTSQHMQGLAADLRCDDPKALFDL---IAESDLPFDQLIYYQKKKFVHVSYSP 120
R VGGV +S H+ GLAAD C D+ I S + FDQLI Q+ +VH+ +P
Sbjct 68 RAVGGVPSSAHLSGLAADFVCPKIGTPLDICRAIRASPIEFDQLI--QEGTWVHIGLAP 124
> cak:Caul_2481 peptidase M15A
Length=146
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 36/85 (42%), Positives = 51/85 (60%), Gaps = 9/85 (11%)
Query 41 LDPVREMYGKPIVVTSGFRTPFLNRLVGGVSTSQHMQGLAADLRCDDPKALFDL-----I 95
++ VR ++ +PI +TS +R P +N VGGV TS H G AAD D + DL +
Sbjct 42 MEAVRALFDRPIEITSAYRNPQVNAAVGGVPTSAHALGHAADFHVD---GVADLDAAKRV 98
Query 96 AESDLPFDQLIYYQKKKFVHVSYSP 120
+S L FDQLI Y+K + VH+S+ P
Sbjct 99 RDSGLKFDQLI-YEKNRCVHISFEP 122
> del:DelCs14_3869 peptidase M15A
Length=151
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 45/122 (37%), Positives = 67/122 (55%), Gaps = 12/122 (10%)
Query 6 YFSFTEMILTASTAQE---NNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
+FS TEM +STAQ +N P + ++RL L +L+ VR G PI+VTSG+R+
Sbjct 6 HFSLTEMT-ASSTAQRQGLDNTPTPEALHRL-ALTAAMLERVRAHLGVPIIVTSGYRSRA 63
Query 63 LNRLVGGVSTSQHMQGLAADLRCDDPKALFDLIAE-----SDLPFDQLIY--YQKKKFVH 115
+N VGGV++S H G AAD+ A +++ + L Q+IY K +VH
Sbjct 64 VNAAVGGVTSSDHAIGAAADIVAPKFGAPYNVAKALAPHVNALGIGQIIYESVGGKHWVH 123
Query 116 VS 117
+S
Sbjct 124 LS 125
> fae:FAES_1814 peptidase M15A
Length=159
Score = 65.5 bits (158), Expect = 2e-11, Method: Compositional matrix adjust.
Identities = 48/140 (34%), Positives = 73/140 (52%), Gaps = 17/140 (12%)
Query 5 KYFSFTEMI--LTASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
K+ + +E I TA+ NN+P + + ++ L + P+ E +GK + V+S FR P
Sbjct 9 KWLTLSEAIKSQTATRKGINNMPTQREYDNMVRLYNAVYVPICERFGK-LPVSSFFRCPA 67
Query 63 LNRLVGGVSTSQHMQGLAADLRCDD-------PKALFDLIAESDLPFDQLIYY-----QK 110
LN+ VGG +TS H+QG A D+ CD K LF+ + + L FDQ+I
Sbjct 68 LNKAVGGSATSDHVQGRAIDIDCDGLGEGYPTNKELFEWV-RTHLEFDQVIQEFPDADGN 126
Query 111 KKFVHVSY-SPTYRHEVIVK 129
+VHV Y SPT + +K
Sbjct 127 PGWVHVGYRSPTQNRKRALK 146
> nmd:NMBG2136_0875 Peptidase M15 family protein
Length=157
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 58/112 (52%), Gaps = 13/112 (12%)
Query 5 KYFSFTEMIL--TASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
K FS E+ TA A N P D++ + + L+ +RE G+PI+VTS FR+
Sbjct 5 KNFSLRELTRSETARRAGVPNEPSPADMDNIYYTAQQ-LEKIREYVGRPIIVTSCFRSER 63
Query 63 LNRLVGGVSTSQHMQGLAADLRCDD--------PKALFDLIAESDLPFDQLI 106
+N+LVGG TS H GLAAD CD K L + E L FDQLI
Sbjct 64 VNKLVGGSPTSAHRHGLAAD--CDASGMTSPAFAKLLIKMRDEGALKFDQLI 113
> nmc:NMC0877 ATP binding protein
Length=157
Score = 63.5 bits (153), Expect = 1e-10, Method: Compositional matrix adjust.
Identities = 45/112 (40%), Positives = 58/112 (52%), Gaps = 13/112 (12%)
Query 5 KYFSFTEMIL--TASTAQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTPF 62
K FS E+ TA A N P D++ + + L+ +RE G+PI+VTS FR+
Sbjct 5 KNFSLRELTRSETARRAGVPNEPSPADMDNIYYTAQQ-LEKIREYVGRPIIVTSCFRSER 63
Query 63 LNRLVGGVSTSQHMQGLAADLRCDD--------PKALFDLIAESDLPFDQLI 106
+N+LVGG TS H GLAAD CD K L + E L FDQLI
Sbjct 64 VNKLVGGSPTSAHRHGLAAD--CDASGMTSPAFAKLLIKMRDEGALKFDQLI 113
> emi:Emin_1075 peptidase M15A
Length=157
Score = 62.8 bits (151), Expect = 2e-10, Method: Compositional matrix adjust.
Identities = 42/126 (33%), Positives = 68/126 (54%), Gaps = 6/126 (5%)
Query 4 PKYFSFTEMILTAST--AQENNLPDWDDINRLLELCRWILDPVREMYGKPIVVTSGFRTP 61
K FSF E+ T + A+ N + + L LC IL+ V +MY K ++TS FR
Sbjct 7 SKNFSFYELAKTDNKNFAESNIIEAAIYKDALYNLCADILERVYDMYAKKPLITSAFRCA 66
Query 62 FLNRLVGGVSTSQHMQGLAADLRCDD--PKALFDLIAESDLPFDQLIY--YQKKKFVHVS 117
LN+++GG SQH++G AAD ++ + +F + S+L + QLI+ + ++H+S
Sbjct 67 DLNKVLGGAPNSQHIKGEAADFILEEVSNEEIFLRLRNSNLFYGQLIWEKFGASSWIHIS 126
Query 118 YSPTYR 123
YR
Sbjct 127 LGYPYR 132
Lambda K H a alpha
0.323 0.139 0.431 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 127182003372