bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-7_CDS_annotation_glimmer3.pl_2_1
Length=354
Score E
Sequences producing significant alignments: (Bits) Value
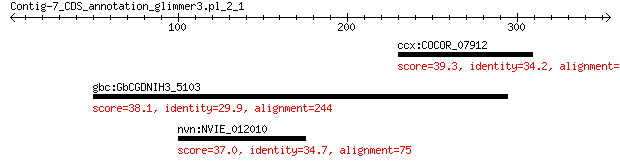
ccx:COCOR_07912 hypothetical protein 39.3 1.3
gbc:GbCGDNIH3_5103 hypothetical protein 38.1 3.2
nvn:NVIE_012010 exported protein of unknown function 37.0 4.2
> ccx:COCOR_07912 hypothetical protein
Length=1121
Score = 39.3 bits (90), Expect = 1.3, Method: Composition-based stats.
Identities = 27/79 (34%), Positives = 41/79 (52%), Gaps = 7/79 (9%)
Query 230 IASRLEIEARTQGQHISNKIARSTADAVIDATCTAKENEAAFNRGYSQFSNDVGFRTGKM 289
+A RL+++ R G + + R AD ++AT AKE EA+ RGYS S +TG
Sbjct 640 LALRLDLDGRVGGIVVDAE-GRPLADVTVEAT--AKEEEASTGRGYSPLSA----KTGPD 692
Query 290 DRWSQDPVKARWDRGINNA 308
R+ +P+ WD + A
Sbjct 693 GRFVLEPLARDWDYELTAA 711
> gbc:GbCGDNIH3_5103 hypothetical protein
Length=1656
Score = 38.1 bits (87), Expect = 3.2, Method: Compositional matrix adjust.
Identities = 73/264 (28%), Positives = 116/264 (44%), Gaps = 46/264 (17%)
Query 50 KSRDF----AKSMFDATNEWNS--AKNQRARLEEAGLNPYLMMNGGSAGTASSTSANTVS 103
K+RD+ AKS D N+ N+ A ++ R +A + +A A S NTVS
Sbjct 489 KNRDYFTSQAKSYVDQQNKLNADIAAAEKKRASDARASRQSATADRAADRADLRSDNTVS 548
Query 104 GASGSGGSPYQFTPTNMIGDVASYAAAMKSMSDARKTNTESDLLDQYGAATYESRIKKTM 163
G S +Q T GD AA+ ++ +K NT++ + G A Y + +K +
Sbjct 549 ALEGQISSLHQLTVAYHEGD-----AAIAGITAQQKANTDALKYGREGTAAYAAEYQKVL 603
Query 164 ADTYFTQRQADVAIAQRANLLLRNDAQEVLNM-YLPEEKRIQLQMNGAQYWNMIREGVIS 222
+ Q VA A++ L + +A + ++ Y+ E R L MN A
Sbjct 604 --PLYQQ----VAEAEQTLALAKKNAADQSSLDYIEAETRT-LGMNDA------------ 644
Query 223 EEQAKNLIASRLEIEARTQGQHISNKIAR---STADAVIDATCTAKENEAAFNR----GY 275
E++K L +LE E R QG + + A+ + D +I+A +AAFN G
Sbjct 645 -ERSKMLSHMKLEAEFRAQGIPLLSDEAQKRIALTDRIIEANGVLARQQAAFNELQSFGE 703
Query 276 SQFSNDVG------FRTGKMDRWS 293
+ F N +G F TGK++ S
Sbjct 704 NTF-NTIGTAITQAFATGKLEAVS 726
> nvn:NVIE_012010 exported protein of unknown function
Length=237
Score = 37.0 bits (84), Expect = 4.2, Method: Compositional matrix adjust.
Identities = 26/75 (35%), Positives = 32/75 (43%), Gaps = 4/75 (5%)
Query 100 NTVSGASGSGGSPYQFTPTNMIGDVASYAAAMKSMSDARKTNTESDLLDQYGAATYESRI 159
N V G G P TPTN I D A A +++++ TN E+ LL Y Y S
Sbjct 144 NVVVGVEPGIGKPSGPTPTNEIIDNAKAVAKSRAVAELNNTNVEAKLLAVY----YNSEF 199
Query 160 KKTMADTYFTQRQAD 174
K A T Q D
Sbjct 200 PKGAAMFVITSEQGD 214
Lambda K H a alpha
0.312 0.126 0.352 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 669457732882