bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_6
Length=168
Score E
Sequences producing significant alignments: (Bits) Value
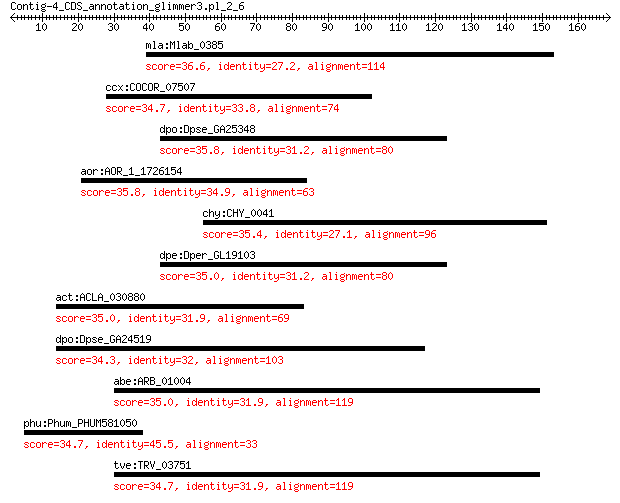
mla:Mlab_0385 hypothetical protein 36.6 2.1
ccx:COCOR_07507 trxC; thioredoxin 34.7 2.9
dpo:Dpse_GA25348 GA25348 gene product from transcript GA25348-RA 35.8 3.5
aor:AOR_1_1726154 AO090003000971; MFS transporter 35.8 3.7
chy:CHY_0041 dnaB; replicative DNA helicase (EC:3.6.1.-) 35.4 4.5
dpe:Dper_GL19103 GL19103 gene product from transcript GL19103-RA 35.0 5.5
act:ACLA_030880 MFS transporter, putative 35.0 6.0
dpo:Dpse_GA24519 GA24519 gene product from transcript GA24519-RA 34.3 8.0
abe:ARB_01004 SNF2 family helicase/ATPase, putative 35.0 8.1
phu:Phum_PHUM581050 Epidermal growth factor receptor precursor... 34.7 8.7
tve:TRV_03751 SNF2 family helicase/ATPase, putative 34.7 9.5
> mla:Mlab_0385 hypothetical protein
Length=624
Score = 36.6 bits (83), Expect = 2.1, Method: Composition-based stats.
Identities = 31/120 (26%), Positives = 58/120 (48%), Gaps = 9/120 (8%)
Query 39 VLEELPTEPFRFEKV-----GTEENEAVRV-RSDVSMLLHAADMAKKYGTGFVKSMIEMH 92
V+E + TE EK+ G E + V R D + A ++ K + +++ +
Sbjct 140 VIETIATEGKNLEKIADYVIGDELPVPIIVPRYDHHIEAAARNLQKYHNLSLAEALFALE 199
Query 93 RPKSSGLQSEMDLMSDAQILDTIKSRHLQSPSELIAWSEYLIDQAKSIEDEASRIALERE 152
S L E+ + S A I I+ RH+ SP ++I + + + AK+I DE + +A +++
Sbjct 200 NVSLSTLSPEV-MESAAAISSEIEKRHMMSPDQIIGANRH--NTAKAISDEVTTLAPQKK 256
> ccx:COCOR_07507 trxC; thioredoxin
Length=119
Score = 34.7 bits (78), Expect = 2.9, Method: Compositional matrix adjust.
Identities = 25/83 (30%), Positives = 40/83 (48%), Gaps = 10/83 (12%)
Query 28 CPICASSEPDIVLEELPTEPFRFEKVGTEENEAVRVRSDV----SMLLH-----AADMAK 78
C I A++EP++ L+EL P R KV E+EA+ R + + LL M++
Sbjct 38 CDIYAAAEPEL-LKELDGAPMRVVKVNAYEHEALATRFGLFGIPTFLLFRNGKLLGKMSQ 96
Query 79 KYGTGFVKSMIEMHRPKSSGLQS 101
YG + +I H P + Q+
Sbjct 97 YYGKPYFLGVIRDHLPGGAKAQA 119
> dpo:Dpse_GA25348 GA25348 gene product from transcript GA25348-RA
Length=419
Score = 35.8 bits (81), Expect = 3.5, Method: Compositional matrix adjust.
Identities = 25/81 (31%), Positives = 41/81 (51%), Gaps = 8/81 (10%)
Query 43 LPTEPFRFEKVGTEENEAVRVRS-DVSMLLHAADMAKKYGTGFVKSMIEMHRPKSSGLQS 101
+P E F+ +G + +R+ + DV + L D+A YG VKS + H LQ
Sbjct 93 VPLEAFKV-ILGYFYSGTIRISTLDVDVTLKVLDLANMYGLVEVKSALSDH------LQE 145
Query 102 EMDLMSDAQILDTIKSRHLQS 122
MD+ + +ILD + HL++
Sbjct 146 HMDVSNVCKILDAARLYHLEN 166
> aor:AOR_1_1726154 AO090003000971; MFS transporter
Length=518
Score = 35.8 bits (81), Expect = 3.7, Method: Composition-based stats.
Identities = 22/66 (33%), Positives = 34/66 (52%), Gaps = 4/66 (6%)
Query 21 PIREVQICPICASSEPDIVLEELPTEPFRFEKVGTEENEAVR---VRSDVSMLLHAADMA 77
P+R++ I IC +EP ++ LP P E VG +NE + + S V+ + A MA
Sbjct 9 PVRQLIILSICRFAEPVVLTSVLPYLPEMIEYVGVPKNEVAKWVGISSAVTSISQAI-MA 67
Query 78 KKYGTG 83
+GT
Sbjct 68 VTWGTA 73
> chy:CHY_0041 dnaB; replicative DNA helicase (EC:3.6.1.-)
Length=443
Score = 35.4 bits (80), Expect = 4.5, Method: Compositional matrix adjust.
Identities = 26/96 (27%), Positives = 48/96 (50%), Gaps = 6/96 (6%)
Query 55 TEENEAVRVRSDVSMLLHAADMAKKYGTGFVKSMIEMHRPKSSGLQSEMDLMSDAQILDT 114
E+ E + + + L D+ + G ++ S++ M P+++ L+S +++ + +L
Sbjct 57 NEKGEPITILTVTEALKQKGDLERAGGVSYIASLLAMAVPEAT-LESHAEIIEEKAVL-- 113
Query 115 IKSRHLQSPSELIAWSEYLIDQAKSIEDEASRIALE 150
RHL SE I Y A+SI +EA R+ LE
Sbjct 114 ---RHLIDLSEKIKRLSYTEKDAESILEEAERLVLE 146
> dpe:Dper_GL19103 GL19103 gene product from transcript GL19103-RA
Length=356
Score = 35.0 bits (79), Expect = 5.5, Method: Compositional matrix adjust.
Identities = 25/81 (31%), Positives = 41/81 (51%), Gaps = 8/81 (10%)
Query 43 LPTEPFRFEKVGTEENEAVRVRS-DVSMLLHAADMAKKYGTGFVKSMIEMHRPKSSGLQS 101
+P E F+ +G + +R+ + DV + L D+A YG VKS + H LQ
Sbjct 93 VPLEAFKV-ILGYFYSGTIRISTLDVDVTLKVLDVANMYGLVEVKSALSDH------LQE 145
Query 102 EMDLMSDAQILDTIKSRHLQS 122
MD+ + +ILD + HL++
Sbjct 146 HMDVSNVCKILDAARLYHLEN 166
> act:ACLA_030880 MFS transporter, putative
Length=503
Score = 35.0 bits (79), Expect = 6.0, Method: Compositional matrix adjust.
Identities = 22/71 (31%), Positives = 32/71 (45%), Gaps = 2/71 (3%)
Query 14 PGYRKCTPIREVQICPICASSEPDIVLEELPTEPFRFEKVGTEENEAVRVRSDVSMLLHA 73
PG P+ ++ I IC +EP + LP P E +G +NE R S + +
Sbjct 2 PGKTPKLPVSQLLILSICRFAEPVVYTSVLPYLPEMIEDIGIPKNEVARWVGITSAVTSS 61
Query 74 --ADMAKKYGT 82
A MA +GT
Sbjct 62 CQAAMAVSWGT 72
> dpo:Dpse_GA24519 GA24519 gene product from transcript GA24519-RA
Length=173
Score = 34.3 bits (77), Expect = 8.0, Method: Compositional matrix adjust.
Identities = 33/109 (30%), Positives = 52/109 (48%), Gaps = 14/109 (13%)
Query 14 PGYRKCTPIR-EVQICPICAS---SEPDIVLEELPTEPFR--FEKVGTEENEAVRVRSDV 67
PG+R+ R E P+C S S V E+P EPF+ E + T + + DV
Sbjct 20 PGHRQVLAKRSEYFRAPLCGSMLESRQREVRLEVPLEPFKAILEYLYTGKLPLSSL--DV 77
Query 68 SMLLHAADMAKKYGTGFVKSMIEMHRPKSSGLQSEMDLMSDAQILDTIK 116
ML+ D+A Y G+V+++I + LQ +M + + IL+ K
Sbjct 78 DMLIDVRDLAHFYCLGYVETLI------TGYLQQKMSVSNVCAILNAAK 120
> abe:ARB_01004 SNF2 family helicase/ATPase, putative
Length=1120
Score = 35.0 bits (79), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 38/143 (27%), Positives = 62/143 (43%), Gaps = 30/143 (21%)
Query 30 ICASSEPDIVLEELPTEPFRFEKVGTEENEAVRVRSDVSML--------LHAADMAKKYG 81
ICA PDI+ P P G E+ A + + S+L +HA + K+
Sbjct 308 ICAHQRPDIINGSYPPTP-----AGLLESRATLIITPPSILKQWQQEIAIHAPGLRVKHY 362
Query 82 TGFVKSMIEMHRPKSSGLQSEMDLMSDAQILDT----------IKSRHLQ------SPSE 125
TG KS ++ R + L S ++ ++L+T SRH + SP
Sbjct 363 TGLKKSKVD-DRELTHDLASYDIVLMTYKVLNTEIYYAEDPPDRPSRHAKQAPKRKSPLM 421
Query 126 LIAWSEYLIDQAKSIEDEASRIA 148
I+W ID+A+ +E ++S+ A
Sbjct 422 QISWWRVCIDEAQMVESQSSKPA 444
> phu:Phum_PHUM581050 Epidermal growth factor receptor precursor,
putative (EC:2.7.10.2)
Length=1323
Score = 34.7 bits (78), Expect = 8.7, Method: Composition-based stats.
Identities = 15/33 (45%), Positives = 20/33 (61%), Gaps = 0/33 (0%)
Query 5 EKNSCTCFGPGYRKCTPIREVQICPICASSEPD 37
E+ + TCFGPG CT + V+ P C SS P+
Sbjct 519 EECASTCFGPGNGNCTKCKHVRDGPFCYSSCPE 551
> tve:TRV_03751 SNF2 family helicase/ATPase, putative
Length=1120
Score = 34.7 bits (78), Expect = 9.5, Method: Compositional matrix adjust.
Identities = 38/143 (27%), Positives = 62/143 (43%), Gaps = 30/143 (21%)
Query 30 ICASSEPDIVLEELPTEPFRFEKVGTEENEAVRVRSDVSML--------LHAADMAKKYG 81
ICA PDI+ P P G E+ A + + S+L +HA + K+
Sbjct 308 ICAHQRPDIINGSYPPTP-----AGLLESRATLIITPPSILKQWQQEIAIHAPGLRVKHY 362
Query 82 TGFVKSMIEMHRPKSSGLQSEMDLMSDAQILDT----------IKSRHLQ------SPSE 125
TG KS ++ R + L S ++ ++L+T SRH + SP
Sbjct 363 TGLKKSKVD-DRELTHDLASYDIVLMTYKVLNTEIYYAEDPPDRPSRHAKQAPKRKSPLM 421
Query 126 LIAWSEYLIDQAKSIEDEASRIA 148
I+W ID+A+ +E ++S+ A
Sbjct 422 QISWWRVCIDEAQMVESQSSKPA 444
Lambda K H a alpha
0.312 0.127 0.359 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 128113138842