bitscore colors: <40, 40-50 , 50-80, 80-200, >200
BLASTP 2.2.30+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Reference for composition-based statistics: Alejandro A. Schaffer,
L. Aravind, Thomas L. Madden, Sergei Shavirin, John L. Spouge, Yuri
I. Wolf, Eugene V. Koonin, and Stephen F. Altschul (2001),
"Improving the accuracy of PSI-BLAST protein database searches with
composition-based statistics and other refinements", Nucleic Acids
Res. 29:2994-3005.
Database: all_orgs
14,240,465 sequences; 5,121,972,263 total letters
Query= Contig-4_CDS_annotation_glimmer3.pl_2_5
Length=638
Score E
Sequences producing significant alignments: (Bits) Value
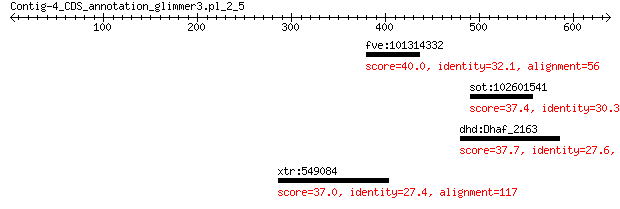
fve:101314332 capsid protein VP1-like 40.0 1.3
sot:102601541 uncharacterized LOC102601541 37.4 4.7
dhd:Dhaf_2163 hypothetical protein 37.7 8.1
xtr:549084 rab43; RAB43, member RAS oncogene family 37.0 8.8
> fve:101314332 capsid protein VP1-like
Length=421
Score = 40.0 bits (92), Expect = 1.3, Method: Compositional matrix adjust.
Identities = 18/56 (32%), Positives = 32/56 (57%), Gaps = 0/56 (0%)
Query 380 LRQAEALQKWKEITQSVDTNYRDQIKAHFGVSVPQSEAHMAKYIGGVARNLDISEV 435
LRQ+ +QK E T Y + I++HFGV+ P + +Y+GG + ++I+ +
Sbjct 311 LRQSFQIQKLLERDARGGTRYTEIIRSHFGVASPDARLQRPEYLGGGSTPINIAPI 366
> sot:102601541 uncharacterized LOC102601541
Length=173
Score = 37.4 bits (85), Expect = 4.7, Method: Compositional matrix adjust.
Identities = 20/66 (30%), Positives = 34/66 (52%), Gaps = 5/66 (8%)
Query 490 SGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVNSADKILGYSPRYYNWKT 549
S PD ++ +ED PIPE DN+ +++ S+ S N +++ + +YYN
Sbjct 47 SEPDSVTPISPLEDFPIPELDNLR-----SLQSQGGSILSETNCSEQDEFVATQYYNELV 101
Query 550 KIDRIH 555
ID+ H
Sbjct 102 SIDKQH 107
> dhd:Dhaf_2163 hypothetical protein
Length=468
Score = 37.7 bits (86), Expect = 8.1, Method: Compositional matrix adjust.
Identities = 29/107 (27%), Positives = 51/107 (48%), Gaps = 3/107 (3%)
Query 480 HAVPLLDYSISGPDGQNLVTSVEDLPIPEFDNIGMESVPAVELMNSSLYSNVNSADKILG 539
H + + +GP G +T E + ++ G + ++ +++Y N S ++LG
Sbjct 52 HKITAVRQEDAGPAGSGPLTLAEMGQLAQYYLNGHDLAYGTKVSATAVYGNNQSLVQVLG 111
Query 540 YSPRYYNWKTKIDRIHGAFTTTLKD--WVAPIDDSFLYSLFGDSLST 584
+ RY + T ID + G+F T + VA ID+ SLFG+ T
Sbjct 112 VNGRYRQFHT-IDLMAGSFLTPGHENQQVAVIDEQLARSLFGNCQVT 157
> xtr:549084 rab43; RAB43, member RAS oncogene family
Length=209
Score = 37.0 bits (84), Expect = 8.8, Method: Composition-based stats.
Identities = 32/130 (25%), Positives = 63/130 (48%), Gaps = 18/130 (14%)
Query 286 GILPNSQFGDLAV-VDLGTISASGSKIPVGAYDGVNDSGTFKQFQTSSESYNGSSDKTTQ 344
G+ Q + V + T+ G ++ + +D ++F+T ++SY S++
Sbjct 38 GVFAERQGSTIGVDFTMKTLEIQGKRVKLQIWDTAGQ----ERFRTITQSYYRSANGAII 93
Query 345 IY-----------PRNPDSISVRADSNLWAVL-GESSDLKLKFSVLALRQAEALQKWKEI 392
Y PR + + A SN+ +L G SDL+ +F +AL++AE L + +I
Sbjct 94 AYDITKRKSFASVPRWIEDVKKYAGSNIVQLLIGNKSDLR-EFREVALQEAETLARHCDI 152
Query 393 TQSVDTNYRD 402
T +++T+ +D
Sbjct 153 TCAIETSAKD 162
Lambda K H a alpha
0.318 0.134 0.414 0.792 4.96
Gapped
Lambda K H a alpha sigma
0.267 0.0410 0.140 1.90 42.6 43.6
Effective search space used: 1497089271516